library(clusterProfiler)
library(org.Hs.eg.db)
library(enrichplot)
library(pathview)
library(ggplotify)
library(ggpubr)GSEA (MOV10)
La hípotesis de GSEA es que cambios pequeños (que no serían detectados por los métodos ORA) pero coordinadosen conjuntos de genes funcionalmente relacionados también pueden tener efectos significativos. En este caso se consideran todos los genes estudiados. Este tipo de análisis puede ser útil en los casos que el análisis de expresión diferencial generé una lista pequeña de genes DE.
Vamos a realizar un análisis de sobrerepresentación con los genes diferencialmente expresados del dataset MOV10 (enlace).
Para realizar el análisis utilizaremos el paquete de Bioconductor clusterProfiler, tambien usaremos el paquete org.Hs.eg.db.
Cargar datos
Cargamos los datos del análisis de expresion diferecial y creamos una lista de los genes ordenados por su FC.
df.fc <- read.csv('../rnaseq/deseq/data/resultado.csv')
df.fc <- df.fc[order(df.fc$log2FoldChange, decreasing = TRUE), ]
geneList <- df.fc$log2FoldChange
names(geneList) <- df.fc$X
head(geneList) HSPA6 MOV10 ASCL1 SCRT1 C1orf95 SNAP25
6.221200 5.082196 4.348903 2.826439 1.902372 1.852436 GO Gene Set Enrichment Analysis
Biological Process
go.BP <- gseGO(geneList = geneList,
OrgDb = org.Hs.eg.db,
keyType = 'SYMBOL',
ont = "BP",
minGSSize = 100,
maxGSSize = 500,
verbose = FALSE)
head(go.BP)| ID | Description | setSize | enrichmentScore | NES | pvalue | p.adjust | qvalue | rank | leading_edge | core_enrichment | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| GO:0042254 | GO:0042254 | ribosome biogenesis | 284 | -0.4475101 | -2.315861 | 0 | 0 | 0 | 4287 | tags=49%, list=28%, signal=36% | CINP/NSUN4/RPS23/NIP7/ERAL1/YBEY/RPS15/RPUSD2/RPL7L1/WDR46/GAR1/SBDS/RPS28/GEMIN4/BRIX1/EIF5/DCAF13/KAT2B/ABCE1/RPP25/SUV39H1/FBL/UTP18/HEATR3/RSL1D1/EIF2A/LTV1/NGDN/TSR1/RAN/NOP10/PA2G4/NOLC1/BYSL/MDN1/MRPS2/C1orf131/NOL8/MPV17L2/IMP3/WDR55/RRP1/FCF1/SURF6/UTP15/NUP88/RPL23A/FTSJ3/REXO4/PWP1/EXOSC3/DKC1/GTPBP10/FDXACB1/CUL4B/NOC4L/PWP2/EIF4A3/XRN2/URB1/SHQ1/RPP30/IMP4/MAK16/RPL26L1/EBNA1BP2/HEATR1/RRS1/RRP15/EXOSC10/DDX28/DDX21/DDX27/XRCC5/WDR43/RRP36/NSUN5/ISG20L2/EIF1AX/DDX47/ABT1/MRPL20/AATF/MPHOSPH10/TRMT112/NOL10/DDX54/NAT10/TSR2/NOC2L/EXOSC5/C1QBP/ERI3/DDX18/PELP1/TRMT2B/POP4/CUL4A/SDAD1/EXOSC6/UTP14A/EIF6/DHX30/NOL11/DDX49/WDR74/PES1/LSG1/NOP14/NOP16/DDX56/KRI1/DDX51/PDCD11/RPS19BP1/NOL9/NGRN/NOL6/MRTO4/GNL3L/SIRT7/NOP2/NHP2/EMG1/RPUSD4/RRP1B/RPSA/RRP7A/NLE1/RRP9/URB2/RPP40/GRWD1/MRPS7/PPAN/MYBBP1A/UTP20/DHX37/RPUSD1 |
| GO:0022613 | GO:0022613 | ribonucleoprotein complex biogenesis | 415 | -0.4336013 | -2.282947 | 0 | 0 | 0 | 4287 | tags=48%, list=28%, signal=36% | CINP/NSUN4/RPS23/NIP7/ERAL1/STRAP/YBEY/RPS15/RPUSD2/RPL7L1/WDR46/GAR1/SBDS/RPS28/GEMIN4/DDX42/BRIX1/SRPK1/EIF5/POLR2D/DCAF13/KAT2B/ABCE1/EIF3J/RPP25/SUV39H1/FBL/UTP18/SNRPD1/EIF3G/HEATR3/RSL1D1/EIF2A/LTV1/NGDN/TSR1/RAN/NOP10/PA2G4/GEMIN7/NOLC1/BYSL/MDN1/MRPS2/C1orf131/CELF1/NOL8/MPV17L2/IMP3/WDR55/RRP1/FCF1/LSM4/HSP90AB1/LSM2/SURF6/SNRPD2/RUVBL2/SNRPG/SF3B2/UTP15/NUP88/CLP1/RPL23A/USP4/FTSJ3/DENR/REXO4/PWP1/EXOSC3/EIF3I/DKC1/GTPBP10/FDXACB1/CUL4B/DICER1/NOC4L/PWP2/EIF4A3/XRN2/LUC7L2/URB1/CD2BP2/SF3A3/SHQ1/PRMT7/RPP30/IMP4/MAK16/HTATSF1/ADAR/RPL26L1/SF1/EBNA1BP2/HEATR1/RRS1/RRP15/EXOSC10/DDX28/SRSF6/DDX21/EIF3A/DDX27/XRCC5/PRPF6/WDR43/SF3B1/RRP36/DDX1/EIF3B/NSUN5/ISG20L2/EIF1AX/PRPF19/SFSWAP/DDX47/ABT1/COIL/SNRNP200/MRPL20/DDX23/NCBP1/AATF/MPHOSPH10/SNRPB/TRMT112/NOL10/DDX54/NAT10/TSR2/NOC2L/EXOSC5/C1QBP/ERI3/U2AF2/TSSC4/DDX18/PELP1/RUVBL1/TRMT2B/POP4/CUL4A/SDAD1/EXOSC6/UTP14A/EIF6/DHX30/NOL11/SNRPD3/DDX49/WDR74/PES1/LSG1/NOP14/NOP16/PUF60/PRMT5/DDX56/KRI1/SRSF9/DDX51/PDCD11/TARBP2/SNRPC/RPS19BP1/SF3B3/NOL9/PTBP2/NGRN/NOL6/MRTO4/PRPF8/GNL3L/SIRT7/PRPF3/NOP2/NHP2/EMG1/CPSF7/RPUSD4/RRP1B/DHX9/WDR77/RPSA/SF3B4/RRP7A/NLE1/RRP9/CELF2/SF3A1/URB2/RPP40/GRWD1/MRPS7/PPAN/MYBBP1A/UTP20/DHX37/RPUSD1 |
| GO:0030198 | GO:0030198 | extracellular matrix organization | 230 | 0.5478588 | 2.276120 | 0 | 0 | 0 | 3599 | tags=47%, list=23%, signal=36% | COL23A1/OLFML2A/ADAMTS7/COL5A1/ADAMTS2/ADAMTS1/COL13A1/DPP4/RECK/CREB3L1/ADAMTS10/LAMC1/ADAMTSL3/EGFLAM/COL2A1/SULF1/THSD4/MMP2/ITGA8/LAMA1/PAPLN/NTN4/NID2/LAMB1/MATN3/HAPLN2/COL3A1/NPNT/LUM/KAZALD1/RAMP2/CSGALNACT1/ABI3BP/SLC2A10/LOXL2/SOX9/LRP1/PDGFRA/ADAMTSL4/COL14A1/PTX3/COL4A4/ADAMTS18/COL27A1/LOXL4/ADAMTS4/OLFML2B/PXDN/COL24A1/MATN2/COL19A1/FBLN1/SMOC2/TGFB1/CAV2/APBB2/LAMB2/COL4A6/COL12A1/CAV1/NDNF/BCL3/ADAMTS3/TNF/ITGB1/GAS6/ADAMTS9/MMP14/WDR72/ADAMTS15/FKBP10/COL4A1/ADAMTS8/SERPINH1/APP/CCDC80/ERCC2/NPHP3/BMP1/FURIN/ADAMTS6/ADAMTS13/FLRT2/MMP15/MMP11/MMP16/FBLN5/PBXIP1/ZNF469/COL4A5/TGFBR3/GPM6B/HMCN1/COL18A1/NID1/HAS3/VPS33B/HAS2/P4HA1/COL4A3/PPARG/COMP/NOTCH1/COL15A1/KLK4/EXOC8/ITGB3 |
| GO:0016072 | GO:0016072 | rRNA metabolic process | 235 | -0.4484916 | -2.267449 | 0 | 0 | 0 | 4222 | tags=47%, list=27%, signal=35% | YBEY/RPS15/RPUSD2/RPL7L1/WDR46/GAR1/SBDS/RPS28/GEMIN4/BRIX1/DCAF13/KAT2B/RPP25/SUV39H1/FBL/UTP18/RSL1D1/NGDN/TSR1/NOP10/PA2G4/NOLC1/BYSL/GTF3C1/NOL8/IMP3/WDR55/RRP1/FCF1/UTP15/POLR1B/FTSJ3/REXO4/PWP1/EXOSC3/GTF3C5/DKC1/FDXACB1/MTOR/NOC4L/PWP2/EIF4A3/XRN2/POLR1E/URB1/SHQ1/RPP30/IMP4/MAK16/EBNA1BP2/HEATR1/RRS1/RRP15/EXOSC10/DDX21/DDX27/WDR43/RRP36/NCL/NSUN5/DDX47/ABT1/SPIN1/MPHOSPH10/TRMT112/NOL10/DDX54/NAT10/TSR2/EXOSC5/ERI3/DDX18/PELP1/TRMT2B/POP4/EXOSC6/UTP14A/SMARCA4/EIF6/NOL11/DDX49/WDR74/PES1/NOP14/GTF3C2/DDX56/KRI1/DDX51/PDCD11/NOL9/NOL6/MRTO4/SIRT7/NOP2/NHP2/EMG1/TCOF1/RPUSD4/RRP1B/BRF1/GTF3C6/RRP7A/DIS3L/POLR2E/RRP9/SMARCB1/RPP40/PPAN/UTP20/DHX37/RPUSD1 |
| GO:0045229 | GO:0045229 | external encapsulating structure organization | 231 | 0.5459670 | 2.266954 | 0 | 0 | 0 | 3599 | tags=46%, list=23%, signal=36% | COL23A1/OLFML2A/ADAMTS7/COL5A1/ADAMTS2/ADAMTS1/COL13A1/DPP4/RECK/CREB3L1/ADAMTS10/LAMC1/ADAMTSL3/EGFLAM/COL2A1/SULF1/THSD4/MMP2/ITGA8/LAMA1/PAPLN/NTN4/NID2/LAMB1/MATN3/HAPLN2/COL3A1/NPNT/LUM/KAZALD1/RAMP2/CSGALNACT1/ABI3BP/SLC2A10/LOXL2/SOX9/LRP1/PDGFRA/ADAMTSL4/COL14A1/PTX3/COL4A4/ADAMTS18/COL27A1/LOXL4/ADAMTS4/OLFML2B/PXDN/COL24A1/MATN2/COL19A1/FBLN1/SMOC2/TGFB1/CAV2/APBB2/LAMB2/COL4A6/COL12A1/CAV1/NDNF/BCL3/ADAMTS3/TNF/ITGB1/GAS6/ADAMTS9/MMP14/WDR72/ADAMTS15/FKBP10/COL4A1/ADAMTS8/SERPINH1/APP/CCDC80/ERCC2/NPHP3/BMP1/FURIN/ADAMTS6/ADAMTS13/FLRT2/MMP15/MMP11/MMP16/FBLN5/PBXIP1/ZNF469/COL4A5/TGFBR3/GPM6B/HMCN1/COL18A1/NID1/HAS3/VPS33B/HAS2/P4HA1/COL4A3/PPARG/COMP/NOTCH1/COL15A1/KLK4/EXOC8/ITGB3 |
| GO:0043062 | GO:0043062 | extracellular structure organization | 231 | 0.5457924 | 2.266229 | 0 | 0 | 0 | 3599 | tags=46%, list=23%, signal=36% | COL23A1/OLFML2A/ADAMTS7/COL5A1/ADAMTS2/ADAMTS1/COL13A1/DPP4/RECK/CREB3L1/ADAMTS10/LAMC1/ADAMTSL3/EGFLAM/COL2A1/SULF1/THSD4/MMP2/ITGA8/LAMA1/PAPLN/NTN4/NID2/LAMB1/MATN3/HAPLN2/COL3A1/NPNT/LUM/KAZALD1/RAMP2/CSGALNACT1/ABI3BP/SLC2A10/LOXL2/SOX9/LRP1/PDGFRA/ADAMTSL4/COL14A1/PTX3/COL4A4/ADAMTS18/COL27A1/LOXL4/ADAMTS4/OLFML2B/PXDN/COL24A1/MATN2/COL19A1/FBLN1/SMOC2/TGFB1/CAV2/APBB2/LAMB2/COL4A6/COL12A1/CAV1/NDNF/BCL3/ADAMTS3/TNF/ITGB1/GAS6/ADAMTS9/MMP14/WDR72/ADAMTS15/FKBP10/COL4A1/ADAMTS8/SERPINH1/APP/CCDC80/ERCC2/NPHP3/BMP1/FURIN/ADAMTS6/ADAMTS13/FLRT2/MMP15/MMP11/MMP16/FBLN5/PBXIP1/ZNF469/COL4A5/TGFBR3/GPM6B/HMCN1/COL18A1/NID1/HAS3/VPS33B/HAS2/P4HA1/COL4A3/PPARG/COMP/NOTCH1/COL15A1/KLK4/EXOC8/ITGB3 |
dotplot(go.BP, showCategory = 15)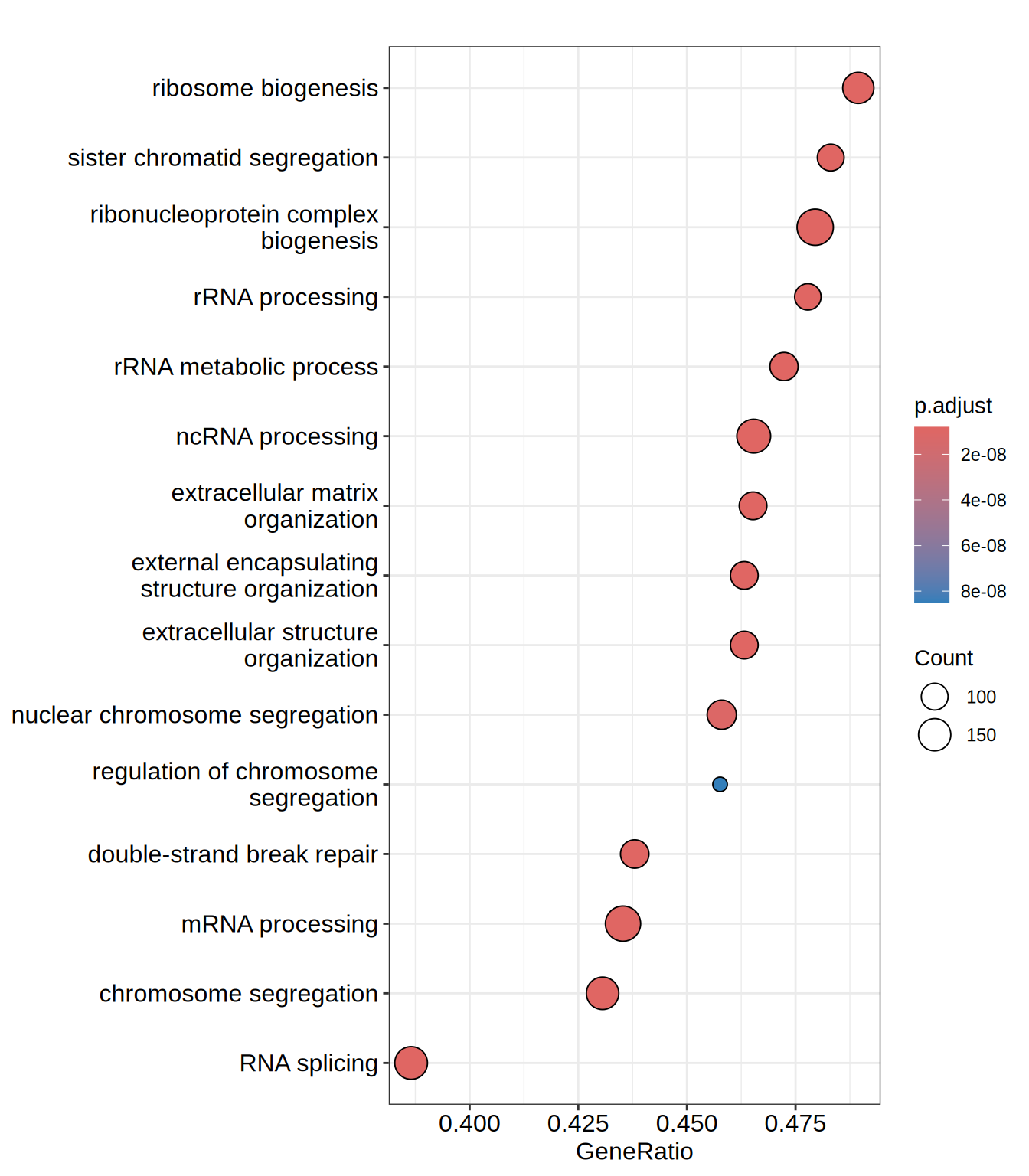
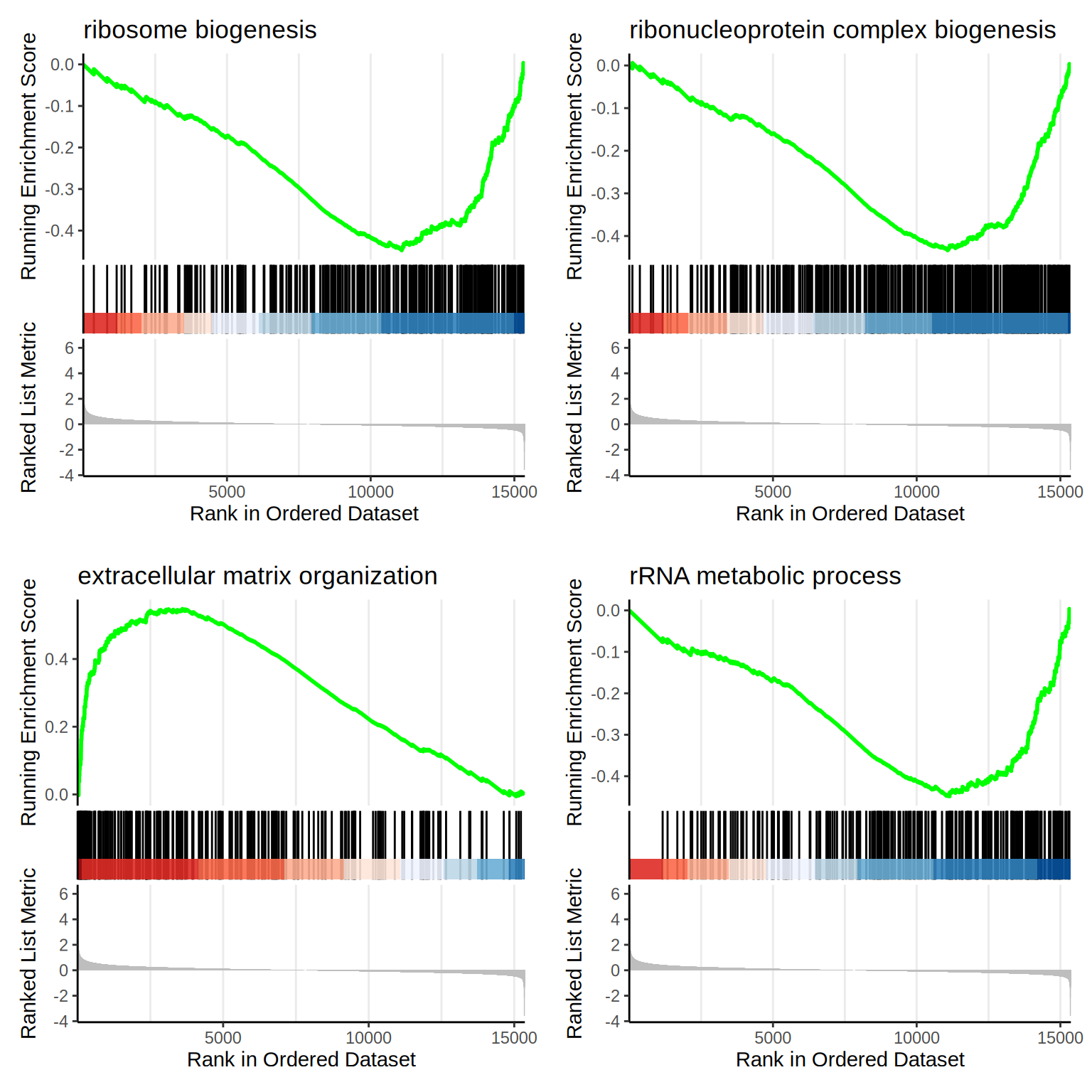
f1 <- as.grob(gseaplot2(go.BP, geneSetID = 1, title = go.BP$Description[1]))
f2 <- as.grob(gseaplot2(go.BP, geneSetID = 2, title = go.BP$Description[2]))
f3 <- as.grob(gseaplot2(go.BP, geneSetID = 3, title = go.BP$Description[3]))
f4 <- as.grob(gseaplot2(go.BP, geneSetID = 4, title = go.BP$Description[4]))
ggarrange(f1, f2, f3, f4,
ncol = 2, nrow = 2
)
Molecular Functions
go.MF <- gseGO(geneList = geneList,
OrgDb = org.Hs.eg.db,
keyType = 'SYMBOL',
ont = "MF",
minGSSize = 100,
maxGSSize = 500,
verbose = FALSE)
head(go.MF)| ID | Description | setSize | enrichmentScore | NES | pvalue | p.adjust | qvalue | rank | leading_edge | core_enrichment | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| GO:0005201 | GO:0005201 | extracellular matrix structural constituent | 105 | 0.6417275 | 2.405642 | 0 | 0e+00 | 0e+00 | 1661 | tags=44%, list=11%, signal=39% | COL23A1/IGFBP7/COL5A1/COL6A2/HSPG2/COL13A1/FBN3/LAMC1/SRPX/ACAN/COL2A1/AGRN/THSD4/LAMA1/NID2/LAMB1/COL21A1/EFEMP1/MATN3/LTBP4/COL3A1/NPNT/LUM/THBS3/ABI3BP/FBN2/COL14A1/LAMA5/COL4A4/EDIL3/COL27A1/CRELD1/LTBP2/EMILIN3/ZP3/PXDN/COL24A1/SPON1/MATN2/COL19A1/FBLN1/LAMB2/COL4A6/LAMA4/COL12A1/VCAN |
| GO:0005509 | GO:0005509 | calcium ion binding | 445 | 0.4432786 | 1.924595 | 0 | 0e+00 | 0e+00 | 3180 | tags=35%, list=21%, signal=28% | SLIT3/DCHS1/CDH23/SCUBE1/HSPG2/CAPN12/SCUBE3/FBN3/RASGRP2/SYT2/EGFLAM/SPOCK1/SYT17/AGRN/SULF1/ITGA5/NOTCH3/DLL1/NID2/GALNT3/EFEMP1/PCDH18/MATN3/LTBP4/MEGF6/SYT6/PCDHB10/DYSF/SNED1/NPNT/CDH3/ITPR3/CDHR1/THBS3/FAM20C/LRP2/NECAB2/EGFL7/HEG1/PLCB2/PCDHB11/LOXL2/TBC1D8B/FBN2/ENPP2/LRP1/KCNIP3/NINL/PAMR1/JAG1/PITPNM1/SCIN/DCHS2/CDH1/EDIL3/FAT4/LRP4/KCNIP2/CRELD1/LTBP2/VLDLR/SYT4/TTYH1/PADI2/PLA2G4A/ANXA1/EFHD1/MATN2/NUCB2/SLC25A25/PROS1/RAPGEF2/FBLN1/PCDH1/SMOC2/FSTL4/PRRG1/CHP2/P4HTM/PCDH10/HSPA5/PCDHB13/ITPR1/PCDHAC2/CDH4/RCN1/CRTAC1/EEF2K/CALML4/VCAN/CLGN/PCDH19/THBD/ANXA4/ENPP3/PLCD1/ITGB1/CDH20/FKBP14/GAS6/CETN3/TRPM4/BAIAP3/PCDH9/PCDH20/UNC13A/MEX3B/GCH1/PAM/RASGRP1/CELSR2/FKBP10/FSTL1/CELSR1/SYT12/ACTN2/S100P/GJB2/FER1L4/EGF/PCDHB14/BMP1/S100A11/ADAMTS13/TBC1D9/PCDHGA11/FAT3/THBS4/LDLR/PCLO/MAN1A1/DSC2/LTBP1/HRC/PPP2R3A/DGKA/FKBP9/FBLN5/CPNE1/EDEM3/PCDHB16/FAT1/NKD2/DNAH7/PCDHB2/HSP90B1/HMCN1/NCALD/NID1/MYL12A/GLCE/LTBP3/IQGAP1/CAPN2/RET |
| GO:0030545 | GO:0030545 | signaling receptor regulator activity | 243 | 0.5066785 | 2.102458 | 0 | 0e+00 | 0e+00 | 2555 | tags=37%, list=17%, signal=31% | CRLF1/INHBE/SCG2/GDNF/MDK/IL11/EGFR/SEMA5B/DPP4/NRTN/GDF15/HSPA1A/TSLP/CXCL12/PGF/DLL1/CHGB/EFEMP1/ADM2/METRNL/IL32/HBEGF/FGF2/CD70/FGF23/FBN2/IL12A/INHBB/CCL20/PDE4D/JAG1/MANF/FLRT3/CSF1/IL18BP/STC1/SEMA4C/TNFSF9/ZP3/PXDN/GDF7/INHBA/SEMA4G/DKK3/FGF5/OSGIN1/FST/TGFB1/TNFSF10/FGF21/CXCL6/CMTM3/CTF1/WNT6/SEMA3E/BMP4/CLEC11A/SEMA6D/TNF/PDGFA/NPPC/GAS6/CSPG5/BMP3/GDF9/VEGFC/PDGFC/WNT11/EDA/LIF/CMTM8/NGF/IL23A/SEMA6B/WNT5B/NPFF/EGF/APP/MRAP2/BMP1/SEMA3D/DKK1/GPNMB/FGF19/HGF/FLRT2/NRG2/FLT3LG/SEMA3F/THBS4 |
| GO:0030546 | GO:0030546 | signaling receptor activator activity | 224 | 0.5079329 | 2.092207 | 0 | 1e-07 | 0e+00 | 2555 | tags=38%, list=17%, signal=32% | CRLF1/INHBE/SCG2/GDNF/MDK/IL11/EGFR/SEMA5B/DPP4/NRTN/GDF15/HSPA1A/TSLP/CXCL12/PGF/DLL1/CHGB/EFEMP1/ADM2/METRNL/IL32/HBEGF/FGF2/CD70/FGF23/FBN2/IL12A/INHBB/CCL20/JAG1/MANF/FLRT3/CSF1/STC1/SEMA4C/TNFSF9/ZP3/GDF7/INHBA/SEMA4G/FGF5/OSGIN1/TGFB1/TNFSF10/FGF21/CXCL6/CMTM3/CTF1/WNT6/SEMA3E/BMP4/CLEC11A/SEMA6D/TNF/PDGFA/NPPC/GAS6/CSPG5/BMP3/GDF9/VEGFC/PDGFC/WNT11/EDA/LIF/CMTM8/NGF/IL23A/SEMA6B/WNT5B/NPFF/EGF/APP/BMP1/SEMA3D/DKK1/GPNMB/FGF19/HGF/FLRT2/NRG2/FLT3LG/SEMA3F/THBS4 |
| GO:0140640 | GO:0140640 | catalytic activity, acting on a nucleic acid | 494 | -0.3168737 | -1.738354 | 0 | 1e-07 | 0e+00 | 3658 | tags=36%, list=24%, signal=28% | LIG1/BRIP1/SMUG1/MLH3/TERC/TSNAX/CHTF8/INO80/POLR2H/RPP14/DHX40/PARN/METTL2B/RAD51/RECQL5/POLA1/ERCC6L/DCP2/RAD51C/DDX12P/RAD54L2/ALKBH1/DALRD3/MCM4/RNASEK/POLR1C/POLR2A/RUVBL2/PGBD5/MSH3/POLR1B/SMARCAD1/MUTYH/QTRT1/FTSJ3/LARS2/POLR3K/EXOSC3/AARS2/THG1L/DKC1/DDX39A/FDXACB1/SUPV3L1/FTO/MCM3/DHX38/METTL6/HELLS/MCM2/DICER1/PCIF1/SAMHD1/EIF4A3/XRCC3/XRN2/PMS2/RBBP8/POLE/FARSB/ALKBH8/RNASEH2A/DFFB/MUS81/FARSA/DHX15/RPP30/TOP1/FEN1/DTWD2/SRCAP/POLR3F/CHRAC1/DBR1/IGHMBP2/MCM7/DNA2/EXOSC10/DDX28/DDX21/DDX27/XRCC5/CHD4/SMARCAL1/ELAC1/DDX1/MLH1/POLR2B/CRCP/NSUN5/POLR2J2/ISG20L2/QRSL1/DDX47/MED20/POLR3B/SNRNP200/DDX23/MSH2/DHX33/ERCC3/TRPT1/DDX54/TOE1/EXOSC5/CNOT7/ERI3/PPP1R8/APEX2/POLR2I/DDX18/POLRMT/RUVBL1/WRNIP1/RAD51D/TRMT2B/POP4/WARS2/DDX19A/RTEL1/PTRHD1/SMARCA4/NTHL1/DHX30/YTHDC2/DDX49/OGG1/POLR3A/ALKBH4/DDX56/DHX16/LCMT2/NSUN2/DDX51/TYW3/POLN/EARS2/POLR1A/GEN1/MAU2/RAD50/XRCC6/ALKBH5/MEPCE/LRRC47/POLR2J/PIF1/POLM/TRMU/DUS3L/TDG/NOP2/EMG1/POP1/DUS1L/DHX9/PUS1/DDX6/DIS3L/POLR2E/POLR2L/TRMT1/CARS2/SMG6/NME1/ANGEL2/RPP40/POLR2F/APEX1/METTL1/DHX34/POLE4/DHX37/TRMT61A/POLR3H/EME2/MSH4 |
| GO:0048018 | GO:0048018 | receptor ligand activity | 219 | 0.5104062 | 2.102871 | 0 | 1e-07 | 1e-07 | 2555 | tags=38%, list=17%, signal=32% | CRLF1/INHBE/SCG2/GDNF/MDK/IL11/SEMA5B/DPP4/NRTN/GDF15/HSPA1A/TSLP/CXCL12/PGF/DLL1/CHGB/EFEMP1/ADM2/METRNL/IL32/HBEGF/FGF2/CD70/FGF23/FBN2/IL12A/INHBB/CCL20/JAG1/MANF/FLRT3/CSF1/STC1/SEMA4C/TNFSF9/ZP3/GDF7/INHBA/SEMA4G/FGF5/OSGIN1/TGFB1/TNFSF10/FGF21/CXCL6/CMTM3/CTF1/WNT6/SEMA3E/BMP4/CLEC11A/SEMA6D/TNF/PDGFA/NPPC/GAS6/CSPG5/BMP3/GDF9/VEGFC/PDGFC/WNT11/EDA/LIF/CMTM8/NGF/IL23A/SEMA6B/WNT5B/NPFF/EGF/APP/BMP1/SEMA3D/DKK1/GPNMB/FGF19/HGF/FLRT2/NRG2/FLT3LG/SEMA3F/THBS4 |
dotplot(go.MF, showCategory = 15)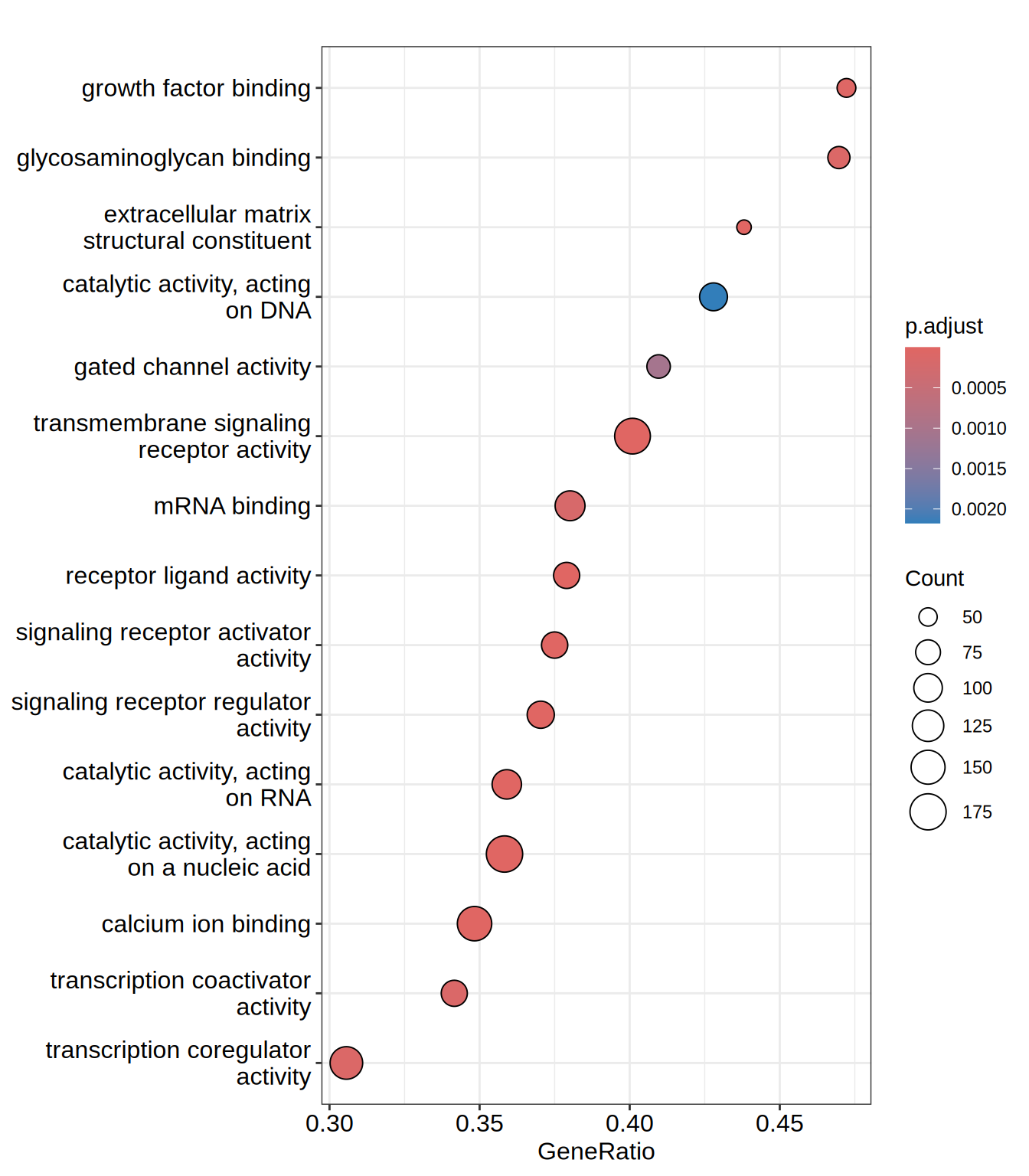
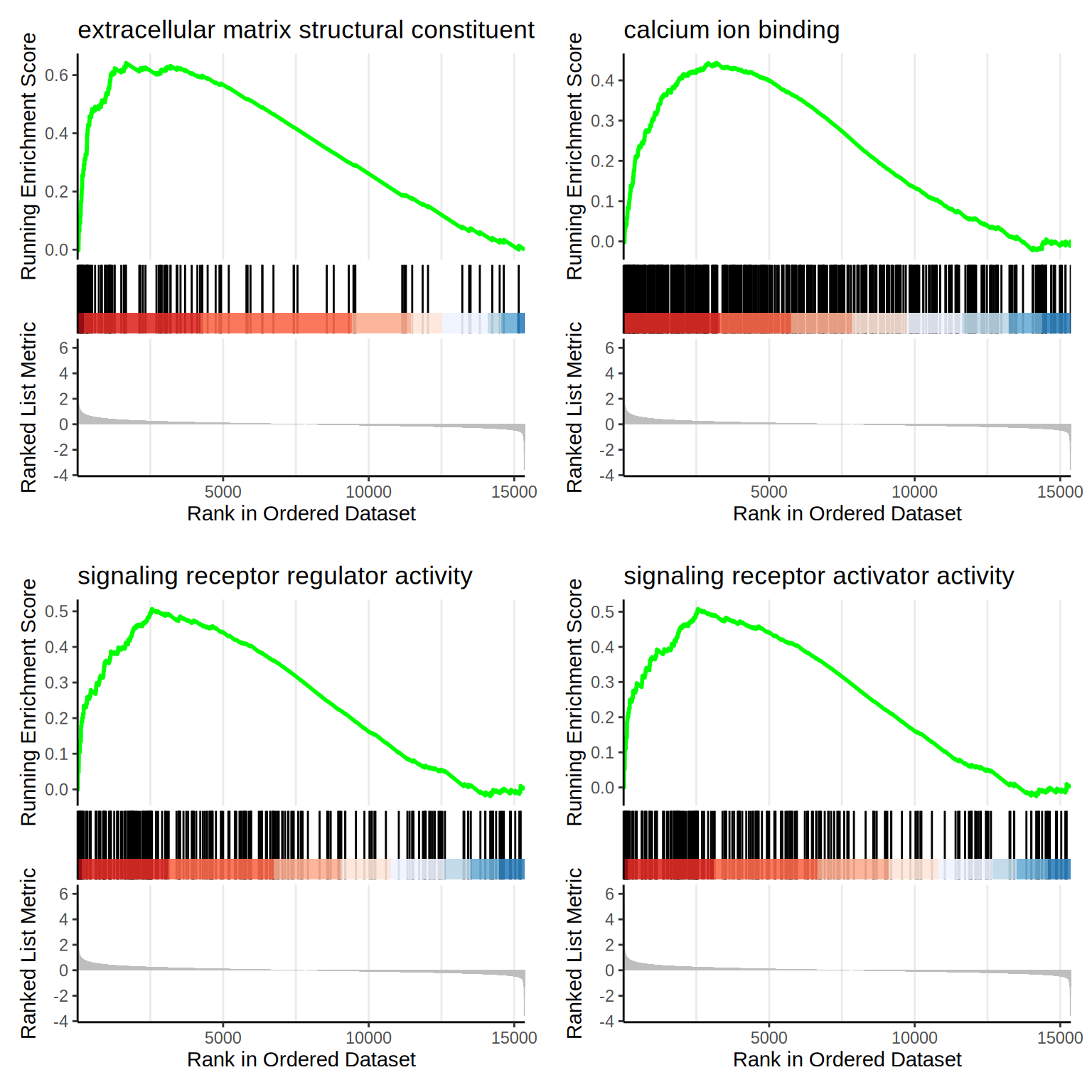
f1 <- as.grob(gseaplot2(go.MF, geneSetID = 1, title = go.MF$Description[1]))
f2 <- as.grob(gseaplot2(go.MF, geneSetID = 2, title = go.MF$Description[2]))
f3 <- as.grob(gseaplot2(go.MF, geneSetID = 3, title = go.MF$Description[3]))
f4 <- as.grob(gseaplot2(go.MF, geneSetID = 4, title = go.MF$Description[4]))
ggarrange(f1, f2, f3, f4,
ncol = 2, nrow = 2
)
Cellular Components
go.CC <- gseGO(geneList = geneList,
OrgDb = org.Hs.eg.db,
keyType = 'SYMBOL',
ont = "CC",
minGSSize = 100,
maxGSSize = 500,
verbose = FALSE)
head(go.CC)| ID | Description | setSize | enrichmentScore | NES | pvalue | p.adjust | qvalue | rank | leading_edge | core_enrichment | |
|---|---|---|---|---|---|---|---|---|---|---|---|
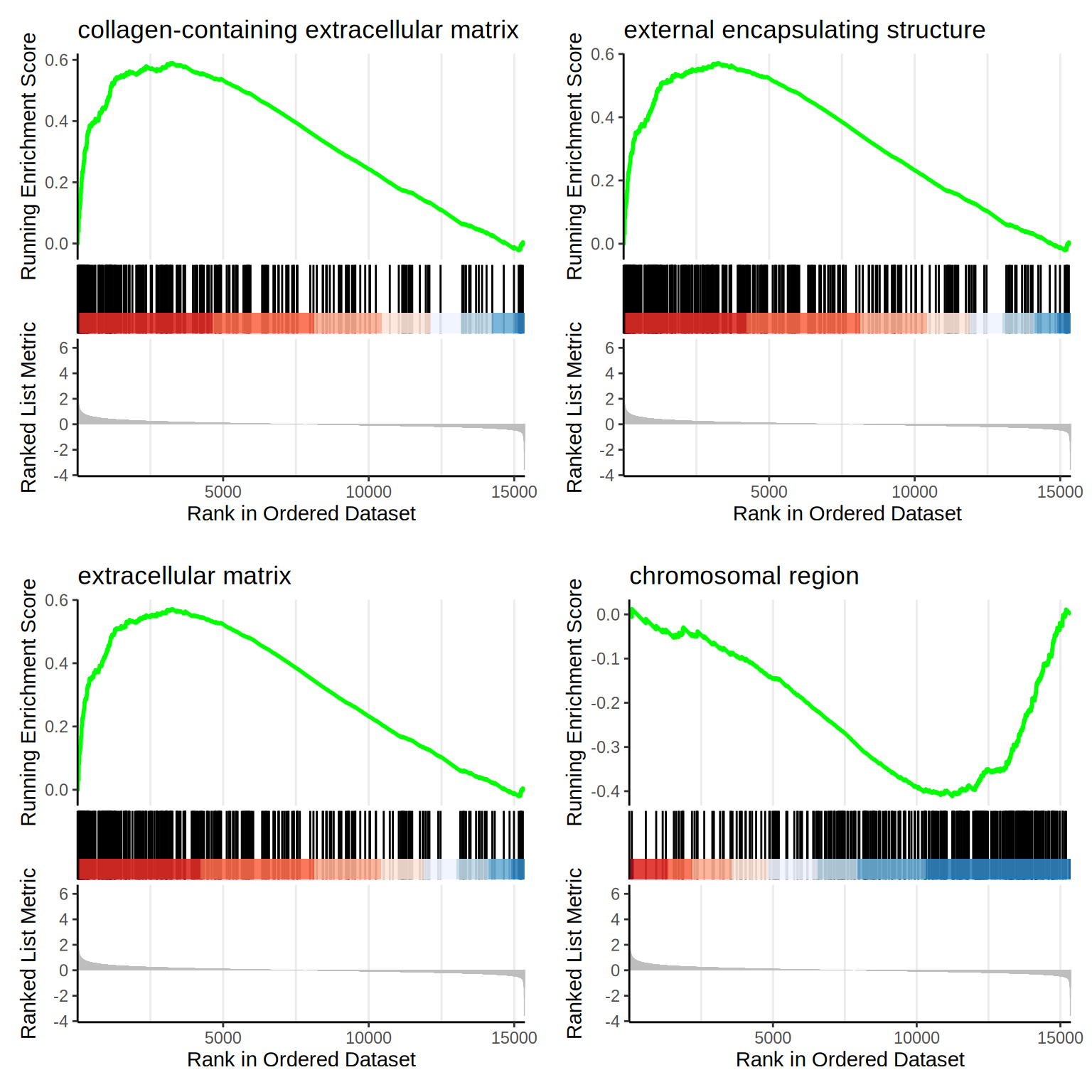
| GO:0062023 | GO:0062023 | collagen-containing extracellular matrix | 259 | 0.5900663 | 2.478964 | 0 | 0 | 0 | 3260 | tags=51%, list=21%, signal=41% | ACHE/INHBE/COL23A1/IGFBP7/MDK/HTRA1/COL5A1/ADAMTS2/COL6A2/ADAMTS1/HSPG2/COL13A1/ITIH5/GDF15/ADAMTS10/LAMC1/SRPX/ACAN/EGFLAM/COL2A1/CXCL12/AGRN/SULF1/THSD4/MMP2/BCAM/LAMA1/NTN4/NID2/LAMB1/COL21A1/EFEMP1/MATN3/LTBP4/HAPLN2/COL3A1/NPNT/LUM/THBS3/KAZALD1/ABI3BP/EGFL7/LOXL2/FBN2/CLU/TIMP3/ADAMTSL4/COL14A1/LAMA5/SERPING1/ACTA2/COL4A4/EDIL3/COL27A1/LOXL4/FREM2/CSPG4/ADAMTS4/CRELD1/LTBP2/EMILIN3/ZP3/PXDN/COCH/GPC6/COL24A1/SPON1/ITGB4/ANXA1/ADAM11/MATN2/L1CAM/COL19A1/FBLN1/SMOC2/CTSC/LAMC3/TGFB1/ADAM19/NAV2/LAMB2/COL4A6/LAMA4/COL12A1/VCAN/SERPINE2/ANXA4/ANXA5/ADAMTS3/GPC5/ADAMTS9/SERPINF1/GPC4/ADAMTS15/FN1/LAMA3/LMAN1/GPC3/COL4A1/WNT5B/HAPLN3/TNC/ADAMTS8/SERPINH1/CCDC80/LGALS3BP/THBS4/LTBP1/APOE/NCAM1/FBLN5/COL4A5/EMILIN2/SFRP1/DCN/HSP90B1/FREM1/MFAP2/C17orf58/HMCN1/COL18A1/SDC3/ANGPTL2/COLEC12/NID1/ANG/LTBP3/MFGE8/COL25A1/FRAS1/GPC1 |
| GO:0030312 | GO:0030312 | external encapsulating structure | 335 | 0.5716858 | 2.446870 | 0 | 0 | 0 | 3260 | tags=49%, list=21%, signal=39% | ACHE/INHBE/COL23A1/OLFML2A/IGFBP7/MDK/ADAMTS7/HTRA1/COL5A1/ADAMTS2/COL6A2/ADAMTS1/HSPG2/COL13A1/ITIH5/GDF15/FBN3/ADAMTS10/LAMC1/SRPX/ACAN/ADAMTSL3/EGFLAM/COL2A1/CXCL12/AGRN/SULF1/THSD4/MMP2/BCAM/LAMA1/NTN4/NID2/LAMB1/COL21A1/EFEMP1/MATN3/LTBP4/HAPLN2/SNED1/COL3A1/NPNT/LUM/THBS3/ELFN1/KAZALD1/ELFN2/ABI3BP/EGFL7/LOXL2/FBN2/CLU/TIMP3/ADAMTSL4/COL14A1/LAMA5/EMID1/SERPING1/ACTA2/PTX3/LRIG3/FLRT3/COL4A4/ADAMTS18/EDIL3/COL27A1/LOXL4/FREM2/CSPG4/ADAMTS4/CRELD1/OLFML2B/LTBP2/EMILIN3/ZP3/PXDN/COCH/GPC6/COL24A1/SPON1/ITGB4/ANXA1/ADAM11/MATN2/OVGP1/L1CAM/COL19A1/FBLN1/LRRN2/SMOC2/CTSC/LAMC3/TGFB1/ADAM19/NAV2/LAMB2/RTN4RL2/COL4A6/LAMA4/LINGO1/COL12A1/WNT6/VCAN/SERPINE2/NDNF/GLG1/ANXA4/ANXA5/ADAMTS3/GPC5/WNT11/ADAMTS9/MMP14/SERPINF1/GPC4/ADAMTS15/LRIG1/FN1/LAMA3/LMAN1/GPC3/COL4A1/WNT5B/HAPLN3/TNC/ADAMTS8/SERPINH1/CCDC80/ADAMTS6/ADAMTS13/FLRT2/LGALS3BP/LINGO3/THBS4/TLR3/MMP15/LTBP1/APOE/NCAM1/MMP11/MMP16/FBLN5/COL4A5/EMILIN2/SFRP1/DCN/HSP90B1/FREM1/MFAP2/C17orf58/HMCN1/COL18A1/FLRT1/SDC3/ANGPTL2/COLEC12/NID1/ANG/LTBP3/MFGE8/COL25A1/TRIL/FRAS1/GPC1 |
| GO:0031012 | GO:0031012 | extracellular matrix | 335 | 0.5716858 | 2.446870 | 0 | 0 | 0 | 3260 | tags=49%, list=21%, signal=39% | ACHE/INHBE/COL23A1/OLFML2A/IGFBP7/MDK/ADAMTS7/HTRA1/COL5A1/ADAMTS2/COL6A2/ADAMTS1/HSPG2/COL13A1/ITIH5/GDF15/FBN3/ADAMTS10/LAMC1/SRPX/ACAN/ADAMTSL3/EGFLAM/COL2A1/CXCL12/AGRN/SULF1/THSD4/MMP2/BCAM/LAMA1/NTN4/NID2/LAMB1/COL21A1/EFEMP1/MATN3/LTBP4/HAPLN2/SNED1/COL3A1/NPNT/LUM/THBS3/ELFN1/KAZALD1/ELFN2/ABI3BP/EGFL7/LOXL2/FBN2/CLU/TIMP3/ADAMTSL4/COL14A1/LAMA5/EMID1/SERPING1/ACTA2/PTX3/LRIG3/FLRT3/COL4A4/ADAMTS18/EDIL3/COL27A1/LOXL4/FREM2/CSPG4/ADAMTS4/CRELD1/OLFML2B/LTBP2/EMILIN3/ZP3/PXDN/COCH/GPC6/COL24A1/SPON1/ITGB4/ANXA1/ADAM11/MATN2/OVGP1/L1CAM/COL19A1/FBLN1/LRRN2/SMOC2/CTSC/LAMC3/TGFB1/ADAM19/NAV2/LAMB2/RTN4RL2/COL4A6/LAMA4/LINGO1/COL12A1/WNT6/VCAN/SERPINE2/NDNF/GLG1/ANXA4/ANXA5/ADAMTS3/GPC5/WNT11/ADAMTS9/MMP14/SERPINF1/GPC4/ADAMTS15/LRIG1/FN1/LAMA3/LMAN1/GPC3/COL4A1/WNT5B/HAPLN3/TNC/ADAMTS8/SERPINH1/CCDC80/ADAMTS6/ADAMTS13/FLRT2/LGALS3BP/LINGO3/THBS4/TLR3/MMP15/LTBP1/APOE/NCAM1/MMP11/MMP16/FBLN5/COL4A5/EMILIN2/SFRP1/DCN/HSP90B1/FREM1/MFAP2/C17orf58/HMCN1/COL18A1/FLRT1/SDC3/ANGPTL2/COLEC12/NID1/ANG/LTBP3/MFGE8/COL25A1/TRIL/FRAS1/GPC1 |
| GO:0098687 | GO:0098687 | chromosomal region | 332 | -0.4116229 | -2.156838 | 0 | 0 | 0 | 4127 | tags=45%, list=27%, signal=33% | GAR1/PPP1R10/HJURP/NDE1/RPA2/SMARCA5/CHMP7/SIN3A/TP53BP1/NUP107/ORC3/KAT2B/SPAG5/SEC13/SUV39H1/GATAD2B/ORC1/MEAF6/AURKB/TERC/NGDN/DCTN3/ZNF618/CHMP3/RAD51/ERCC6L/TEX14/NSL1/DYNC1LI1/DCTN1/TRAPPC12/BOD1/SMARCE1/CDCA8/ZWILCH/MCM4/FBXW11/CENPT/THOC6/PHF2/FBXO28/THOC1/PLK1/ANAPC16/WDR82/EID3/DAXX/PPP2CA/BRD7/PDS5A/MCM3/HELLS/MCM2/HSF1/XRCC3/SUMO3/THOC7/SMC3/CBX1/MTBP/SIRT2/RCC2/ARID2/SPDYA/CDC20/FEN1/ACD/CENPV/CHRAC1/FMR1/NUP98/KIF2C/MCM7/DNA2/SEH1L/HNRNPU/KIF22/XRCC5/SMARCD1/NUP85/CHD4/SMC1A/ZWINT/BAZ1B/POLR2B/SNAI1/NBN/SPC24/FLYWCH1/LRWD1/EZH1/ACTL6A/SPC25/RANGAP1/MSH2/DCTN4/CENPN/CHMP4A/SKA1/NAT10/KDM4A/INCENP/MTA2/WRNIP1/CENPB/RAD51D/PMF1/RTEL1/RPA1/SMARCA4/CENPM/CHMP4B/MEN1/ZW10/SIRT6/C9orf78/DCTN5/PBRM1/TELO2/TFIP11/BIRC5/PARP1/CCNB1/PPP2R1A/PSEN2/RPA4/RAD50/PHF6/STAG1/XRCC6/SKA3/PIF1/DDB1/PPP1CA/CENPI/KAT5/ALYREF/NHP2/CTCF/SMARCC1/CDCA5/CHAMP1/CHMP1A/SMG6/SMARCB1/CHMP6/ACTB/APEX1 |
| GO:0098798 | GO:0098798 | mitochondrial protein-containing complex | 238 | -0.4230790 | -2.114767 | 0 | 0 | 0 | 5011 | tags=53%, list=33%, signal=37% | TIMM8B/UQCRQ/MRPL55/UQCRH/NDUFV1/MRPL2/PHB2/MRPL18/FOXRED1/MRPL43/MRPS22/PAM16/MRPS24/MRPL16/MRPS31/MRPL24/TIMM44/COX5A/CYC1/MRPL30/PDHX/MRPL3/NDUFC1/NSUN4/PMPCA/TIMM17B/MRPL9/NDUFA1/TOMM5/MRPS15/MRPL48/NDUFB9/COX5B/IDH3G/GRPEL1/MRPL51/TIMM22/NDUFV2/NDUFB7/NFS1/MRPS26/MRPS18A/NDUFB10/NDUFA3/MRPL47/MRPS2/SDHC/COX7C/NDUFAB1/MPV17L2/PDHA2/NDUFA11/MRPL40/ISCU/AURKAIP1/MRPS18B/MRPS23/DLD/TOMM20/MRPL46/VDAC1/MRPL49/UQCR11/NDUFA12/MCCC2/MRPS10/SUPV3L1/NDUFB11/AGK/NDUFC2/MRPL52/MRPL44/MRPL14/TIMM13/NDUFS8/PDHA1/MRPL15/MRPL38/NDUFS5/MRPL37/MRPS16/TIMM23/NDUFB8/MRPL53/DNA2/MRPL28/SAMM50/DLAT/MTX1/COX6B1/LYRM4/MRPL45/MRPL1/MRPS12/MRPL41/MRPL12/MRPL20/NDUFA8/TIMM50/NDUFS3/NDUFS2/PPIF/POLRMT/MRPL35/MRPS11/BCS1L/MRPL11/MRPS35/TOMM22/FXN/MRPL23/MRPL4/PDK2/NDUFS6/TOMM40L/TOMM6/IDH3B/MRPL54/NDUFB2/GADD45GIP1/HSD17B10/BAX/DNAJC11/MRPL27/MRPS7/TOMM40/TIMM10 |
| GO:0009897 | GO:0009897 | external side of plasma membrane | 200 | 0.5379539 | 2.209822 | 0 | 0 | 0 | 3218 | tags=47%, list=21%, signal=38% | CRLF1/FCGRT/SCUBE1/PLAU/TNFRSF9/TNFRSF13C/ACE/ABCA1/CXCL12/CD24/ITGA5/BCAM/ATP1B2/ITGA8/ITGA11/ULBP2/IL1R1/MCAM/ITGA4/LRP2/OSMR/ITGA9/MR1/HEG1/S1PR1/MICB/ULBP1/FAS/HLA-B/PDGFRA/FCRLB/ADAM21/CNTFR/GFRA1/ADAM9/NT5E/ITGA6/BTNL9/LEPR/CCR10/BTN3A3/BTN2A2/BTN3A2/TGFBR2/HLA-F/ITGA3/BTN2A3P/CD1D/IL27RA/RTN4RL2/ENPEP/BTN3A1/KCNJ3/IL6R/EPHA5/CLEC4F/CHRNA4/IL4R/THBD/ENPP3/ANXA5/ITGA1/TNF/ITGB1/CX3CR1/SLC2A4/AQP4/SDC1/ITGAL/INSR/ITGA7/KIT/GPC4/GHR/ADA/EPOR/IGSF21/ABCG1/LDLR/HLA-E/NCAM1/HLA-C/IL2RG/ALCAM/TGFBR3/B2M/CXCR4/SLC38A1/LIFR/CD14/CAPN2/IL11RA/MFGE8/RAET1G |
dotplot(go.CC, showCategory = 15)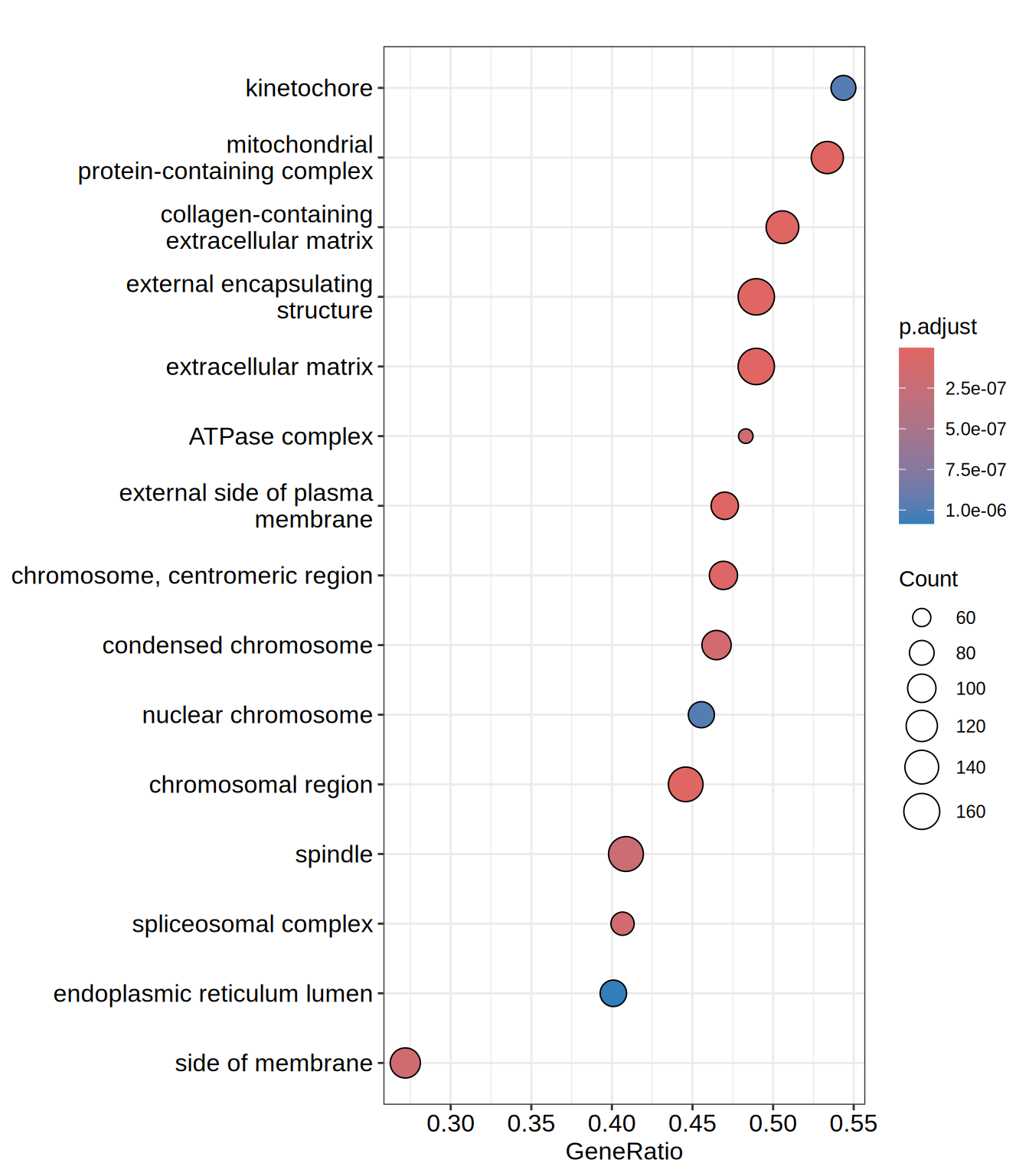
f1 <- as.grob(gseaplot2(go.CC, geneSetID = 1, title = go.CC$Description[1]))
f2 <- as.grob(gseaplot2(go.CC, geneSetID = 2, title = go.CC$Description[2]))
f3 <- as.grob(gseaplot2(go.CC, geneSetID = 3, title = go.CC$Description[3]))
f4 <- as.grob(gseaplot2(go.CC, geneSetID = 4, title = go.CC$Description[4]))
ggarrange(f1, f2, f3, f4,
ncol = 2, nrow = 2
)
KEGG pathway gene set enrichment analysis
Primero debemos de obtener los valores de ENTREZID de los genes.
allgenes.entrezid <- bitr(names(geneList),
fromType = 'SYMBOL',
toType = 'ENTREZID',
OrgDb = 'org.Hs.eg.db'
)
geneList.kegg <- geneList
names(geneList.kegg) <- allgenes.entrezid$ENTREZID
head(geneList.kegg) 3310 4343 429 83482 6616 43
6.221200 5.082196 4.348903 2.826439 1.902372 1.852436 gsea.kegg <- gseKEGG(geneList = geneList.kegg,
organism = 'hsa',
minGSSize = 120,
verbose = FALSE)
head(gsea.kegg)| ID | Description | setSize | enrichmentScore | NES | pvalue | p.adjust | qvalue | rank | leading_edge | core_enrichment | |
|---|---|---|---|---|---|---|---|---|---|---|---|
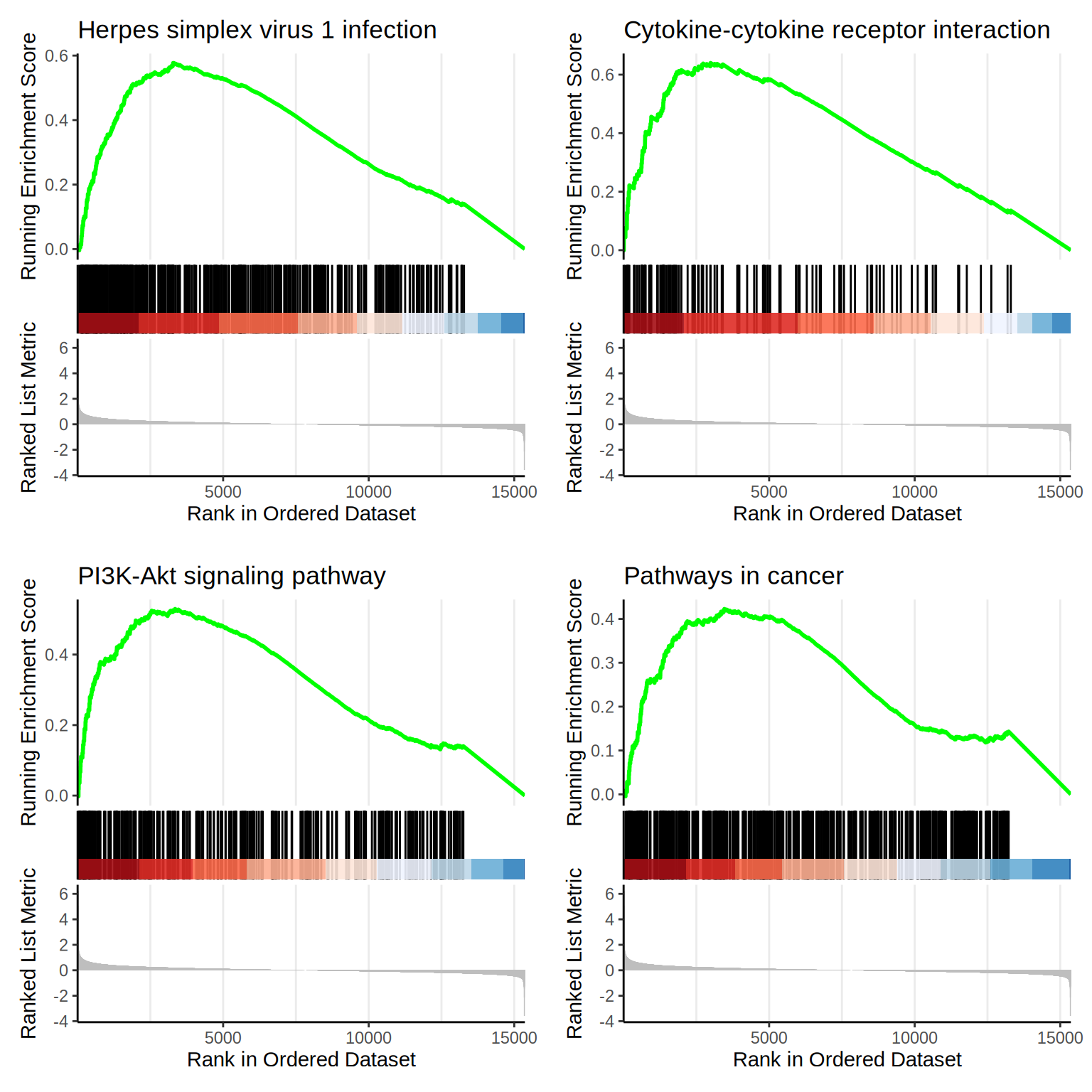
| hsa05168 | hsa05168 | Herpes simplex virus 1 infection | 428 | 0.5772203 | 2.501456 | 0 | 0e+00 | 0e+00 | 3306 | tags=46%, list=22%, signal=37% | 7738/10172/148268/147686/155054/163059/51276/199704/152687/147837/79973/3678/684/7539/126295/342892/55659/10520/10781/199692/10794/284307/388558/9451/163081/7561/90594/148254/10308/148266/147687/10795/9534/7673/7637/147923/342909/355/3106/3592/729747/64135/57573/92283/9422/284370/390980/126231/80778/168374/100131827/374900/93474/7188/84765/7638/140612/84436/57711/5293/163227/84874/79744/57232/3134/162972/7733/201514/7752/162655/339500/92285/90075/55769/117608/7594/80095/79862/162962/374899/7189/58500/146198/7700/342908/7565/1147/9668/7768/84914/163131/55552/10224/374879/148103/7554/84911/54811/90589/7770/7124/84503/65251/284349/57677/83744/7633/219749/7711/388536/169841/162993/7766/163255/90592/84527/285268/9831/163049/340252/7556/7699/400713/10793/126070/147949/163087/284371/728927/7773/348327/7644/51710/125893/374928/163051/94039/8892/7695/284390/170959/3133/7098/147694/6892/7639/91120/284309/3459/3107/79724/29915/148203/58492/567/6432/54807/170960/22835/390927/146434/51427/7692/90321/388566/80110/7574/7767/3105/55713/126375/7576/127396/170958/80264/3690/84924/126068/9310/57547/90576/26974/641339/8717/8772/6428/9021/59348/91664/330/353355/349075/125919/91975/4792 |
| hsa04060 | hsa04060 | Cytokine-cytokine receptor interaction | 124 | 0.6400572 | 2.475342 | 0 | 0e+00 | 0e+00 | 2988 | tags=51%, list=19%, signal=41% | 83729/3589/9518/3604/115650/85480/6387/130399/3554/9235/9180/970/355/3592/3625/1271/6364/3953/51330/60401/2826/1435/27242/8744/7048/151449/3624/9466/7040/8743/8809/3570/3588/6372/8795/1489/3566/652/7124/1524/651/2661/3601/1896/3976/8793/2690/4803/51561/2057/55540/3459/3561/84818/768211/7852/2920/7066/3977/3590/3587/2660/2919 |
| hsa04151 | hsa04151 | PI3K-Akt signaling pathway | 276 | 0.5293662 | 2.214544 | 0 | 0e+00 | 0e+00 | 3349 | tags=40%, list=22%, signal=32% | 2668/5105/1956/1292/4902/90993/3915/1280/5228/1026/3678/8516/284217/22801/54541/3912/64764/1942/3718/2065/9863/595/7059/3676/9180/10161/3680/2247/8074/4609/4602/5156/3655/3911/9586/1943/2786/1435/1286/5293/5106/3691/3675/10319/2250/3913/2792/26291/3570/1288/1147/3910/3566/23533/5879/3672/5154/3688/2149/7424/56034/6198/3643/3679/3815/2690/3696/2335/4803/3909/1282/5894/29941/2057/1950/3371/9965/3082/2323/7060/23566/5563/4908/5523/3561/1287/3693/118788/7184/55970/1969/3667/23035/5979/6655/148327/1285/1311/3690/2321/2788/9170/284/1284/2064/55012/2784/894/1021/2322 |
| hsa05200 | hsa05200 | Pathways in cancer | 436 | 0.4227458 | 1.834197 | 0 | 1e-07 | 0e+00 | 3462 | tags=32%, list=23%, signal=25% | 1956/10235/3915/6387/5228/1026/4313/4854/284217/28514/3912/388585/624/3718/595/5366/1647/10161/840/2247/1643/8074/5330/4609/2735/5915/8322/355/5156/3592/113/3655/3911/1910/182/2786/3725/1286/7188/999/111/5293/7048/3675/816/10319/2250/7040/7189/5292/3913/2792/185/26291/3570/1288/1147/3910/1030/91860/3566/7475/1164/9252/652/5879/5154/3688/23604/2149/7424/5337/7481/3601/6198/3815/10125/2335/1163/3909/51561/1282/81029/5894/2057/6093/1950/4040/1909/3280/9965/3082/4843/3162/2323/1499/23566/3459/3561/1287/7184/55970/7709/6932/7852/5979/3091/6655/112/5338/7855/5732/54361/1285/5468/10023/4851/8326/2788/7043/9170/1284/115/8772/2064/11186/330/2784/4792/894/1021/2322/6667/7473/89780/7422/23493/6199/5336 |
| hsa04820 | hsa04820 | Cytoskeleton in muscle cells | 176 | 0.5244601 | 2.101264 | 0 | 1e-07 | 0e+00 | 3175 | tags=38%, list=21%, signal=31% | 1289/1292/3339/84467/25802/84033/375790/3678/482/8516/284217/22801/2027/22795/2318/1281/7059/3676/3680/2201/3655/287/1286/85301/255631/3691/7138/8572/3675/2192/271/8910/6385/1288/1462/3672/3688/6382/3679/23345/3696/2335/88/1282/23500/6640/7060/114793/1824/1466/27063/1287/3693/1634/9672/4811/1285/1730/1311/25777/7414/23224/6641/3690/4001/752/1284 |
| hsa04080 | hsa04080 | Neuroactive ligand-receptor interaction | 152 | 0.5312897 | 2.099922 | 0 | 2e-07 | 1e-07 | 4074 | tags=53%, list=27%, signal=39% | 1394/117/2696/22953/11255/624/79924/6752/10161/53637/1901/3352/3953/5028/1910/55584/152/10316/344838/79957/1132/185/116444/5029/706/1137/154/2901/2149/153/2690/2906/2899/8620/84109/1909/2743/23566/2894/135/1136/5745/51083/3269/2914/7434/5732/1815/136/10886/2796/2563/55879/4985/10203/2892/89832/9170/2900/2560/10424/797/552/2557/1902/6863/799/27346/7349/59340/5027/5443/3363/6019/7425/1903/6013/4922/887/1143 |
dotplot(gsea.kegg, showCategory = 15)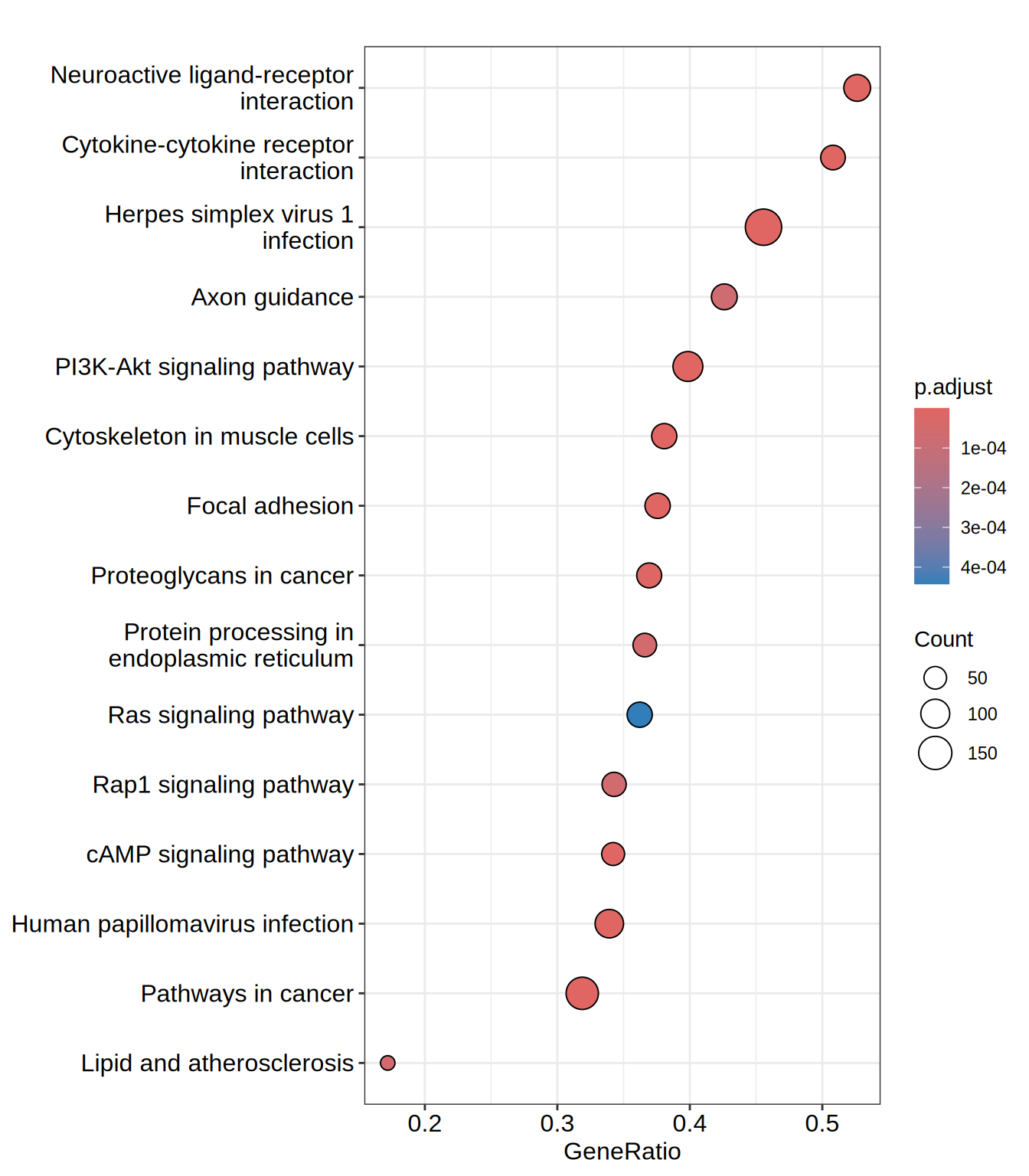
f1 <- as.grob(gseaplot2(gsea.kegg, geneSetID = 1, title = gsea.kegg$Description[1]))
f2 <- as.grob(gseaplot2(gsea.kegg, geneSetID = 2, title = gsea.kegg$Description[2]))
f3 <- as.grob(gseaplot2(gsea.kegg, geneSetID = 3, title = gsea.kegg$Description[3]))
f4 <- as.grob(gseaplot2(gsea.kegg, geneSetID = 4, title = gsea.kegg$Description[4]))
ggarrange(f1, f2, f3, f4,
ncol = 2, nrow = 2
)
Pathways
core <- geneInCategory(gsea.kegg)[['hsa05168']]
hsa05168 <- pathview(gene.data = geneList.kegg,
pathway.id = "hsa05168",
species = "hsa",
limit = list(gene=round(max(abs(geneList.kegg[core])),1), cpd=1))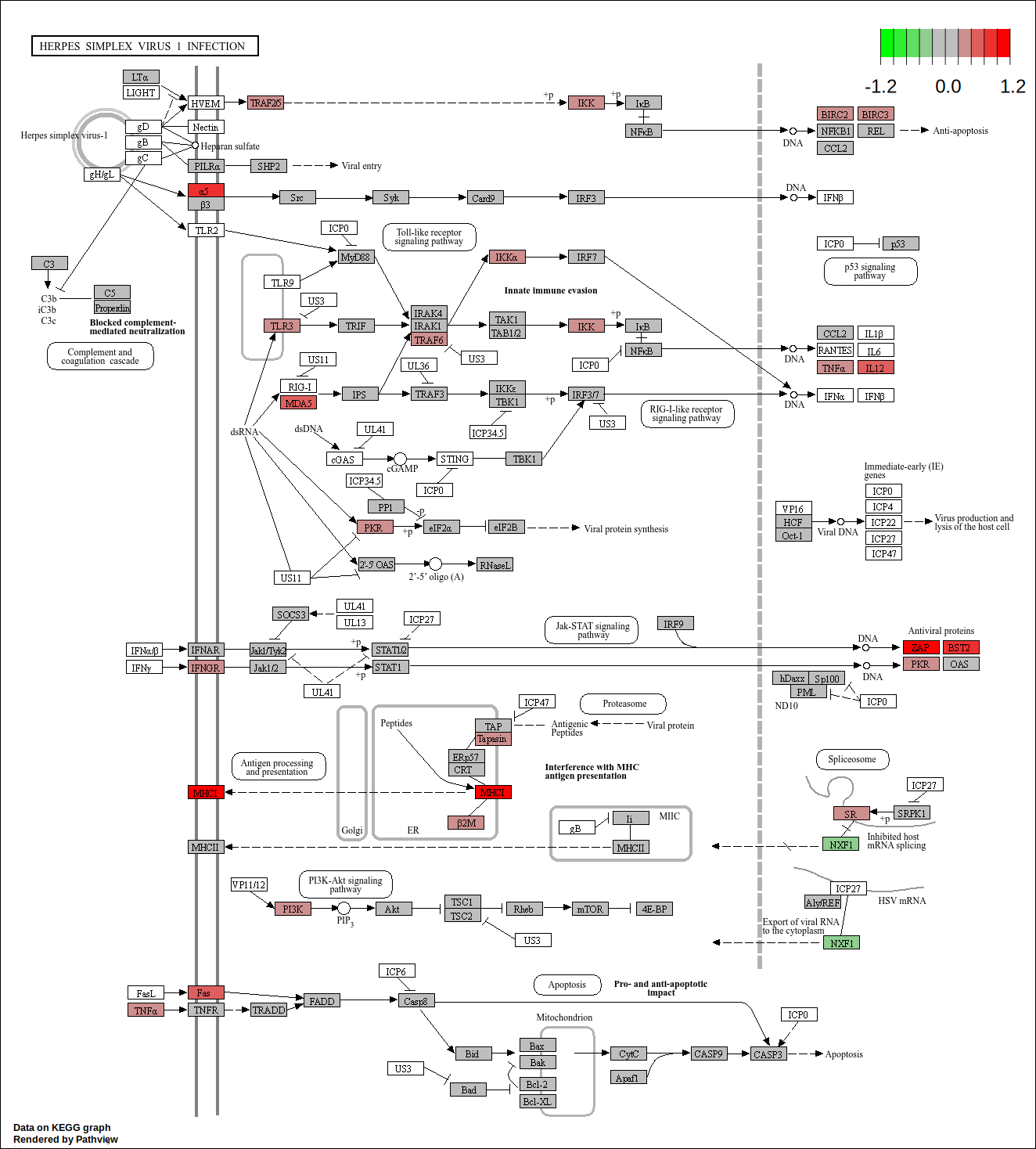
core <- geneInCategory(gsea.kegg)[['hsa04060']]
hsa04060 <- pathview(gene.data = geneList.kegg,
pathway.id = "hsa04060",
species = "hsa",
limit = list(gene=round(max(abs(geneList.kegg[core])), 1), cpd=1))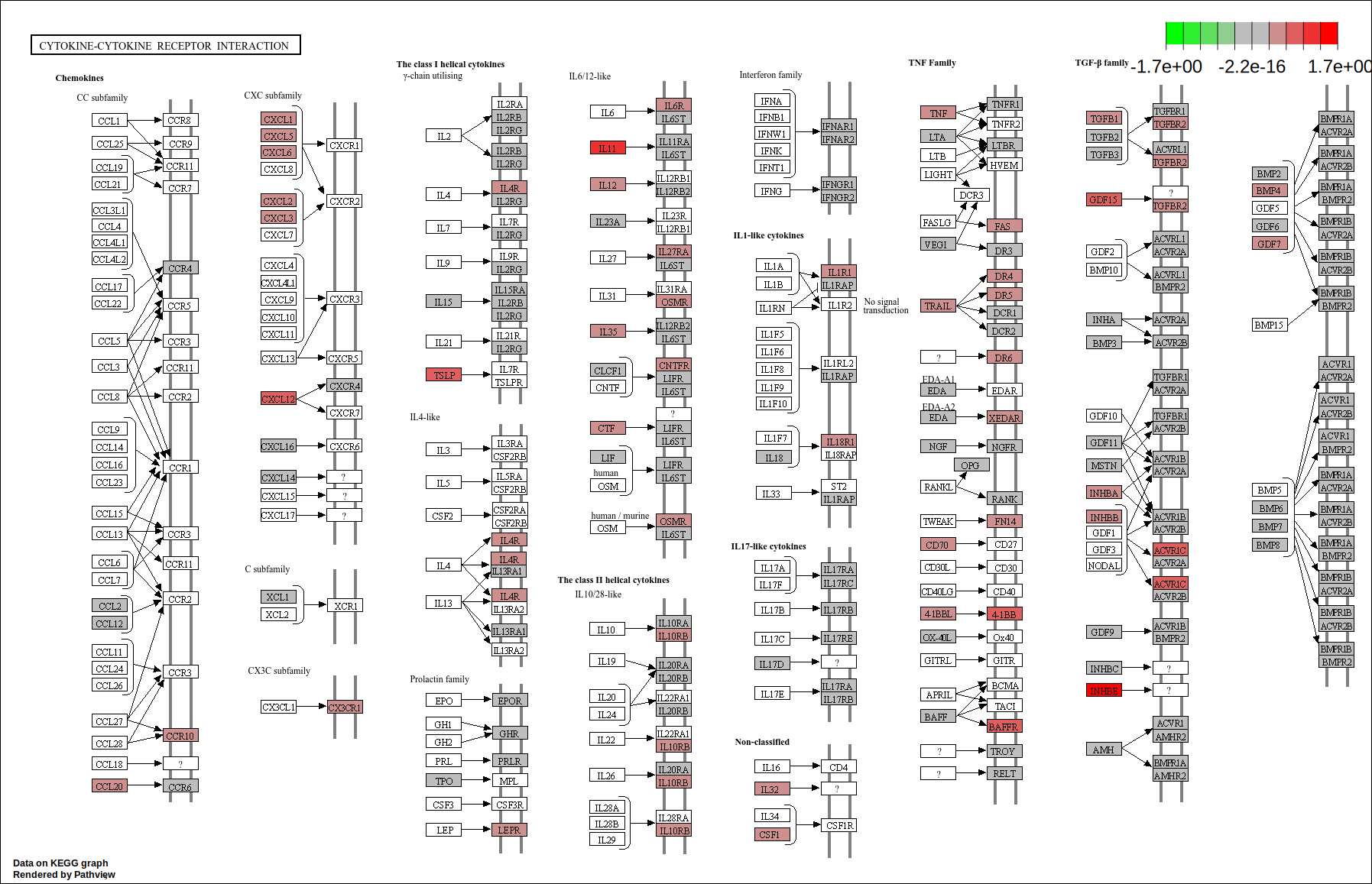
Referencias
citation("clusterProfiler")Please cite S. Xu (2024) for using clusterProfiler. In addition, please
cite G. Yu (2010) when using GOSemSim, G. Yu (2015) when using DOSE and
G. Yu (2015) when using ChIPseeker.
S Xu, E Hu, Y Cai, Z Xie, X Luo, L Zhan, W Tang, Q Wang, B Liu, R
Wang, W Xie, T Wu, L Xie, G Yu. Using clusterProfiler to characterize
multiomics data. Nature Protocols. 2024,
doi:10.1038/s41596-024-01020-z
T Wu, E Hu, S Xu, M Chen, P Guo, Z Dai, T Feng, L Zhou, W Tang, L
Zhan, X Fu, S Liu, X Bo, and G Yu. clusterProfiler 4.0: A universal
enrichment tool for interpreting omics data. The Innovation. 2021,
2(3):100141
Guangchuang Yu, Li-Gen Wang, Yanyan Han and Qing-Yu He.
clusterProfiler: an R package for comparing biological themes among
gene clusters. OMICS: A Journal of Integrative Biology 2012,
16(5):284-287
To see these entries in BibTeX format, use 'print(<citation>,
bibtex=TRUE)', 'toBibtex(.)', or set
'options(citation.bibtex.max=999)'.