ggplot2
Elegant Graphics for Data Analysis
¿Qué es ggplot2?
ggplot2 es un paquete de R para producir visualizaciones de datos. A diferencia de otros paquetes graficos, ggplot2 usa un framework conceptual basado en la gramática de los gráficos. Esto permite crear gráficos de diferentes elementos, en lugar de estar limitados a un conjunto de gráficos predeterminados. A día de hoy es el lider indiscutible en representaciones gráficas en R.
Estructura
Existen 7 elementos que se unen como un conjunto de instrucciones para dibujar un gráfico. Un gráfico presenta al menos tres elementos: Data, mapping, layer.
Estructura
Data: Los datos que se van a representar, generalmente será un data frame.
Mappings:Características estéticas (aes) que describiran cómo queremos que los datos se vean en el gráfico (posición, color, relleno, forma, tamaño, etc..).
Layers: Es la capa que muestra como aparecen los datos. Cada una de ellas tiene 3 partes importantes:
- Geometría: (geom) Determina como se muestra cada punto, linea, etc..
- Transformación estadisticas: (stat) Que puede computar nuevas variables de los datos.
- Ajuste de posición: Determina donde se muestra cada parte de datos.
Instalación
El paquete ggplot2 no se encuentra en R-base, para su uso es necesario descargarlo e instalarlo desde los repositorios de CRAN.
Para poder usarlo hay que cargarlo en el sistema usando la función library()
Diamonds
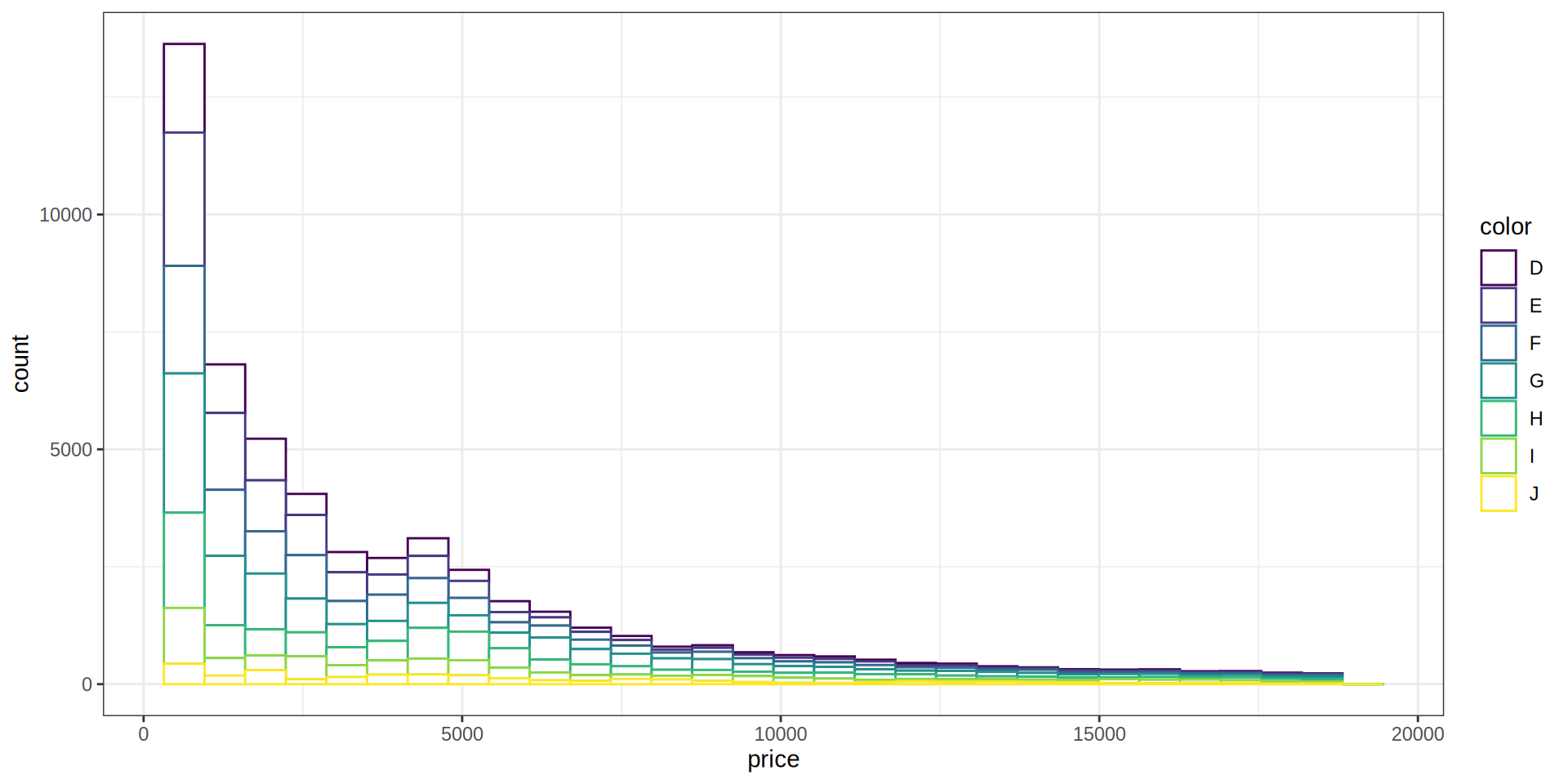
En el paquete ggplot2 esta incluido el dataset Diamonds que contiene el precio y otros atributos de 54.000 diamantes.
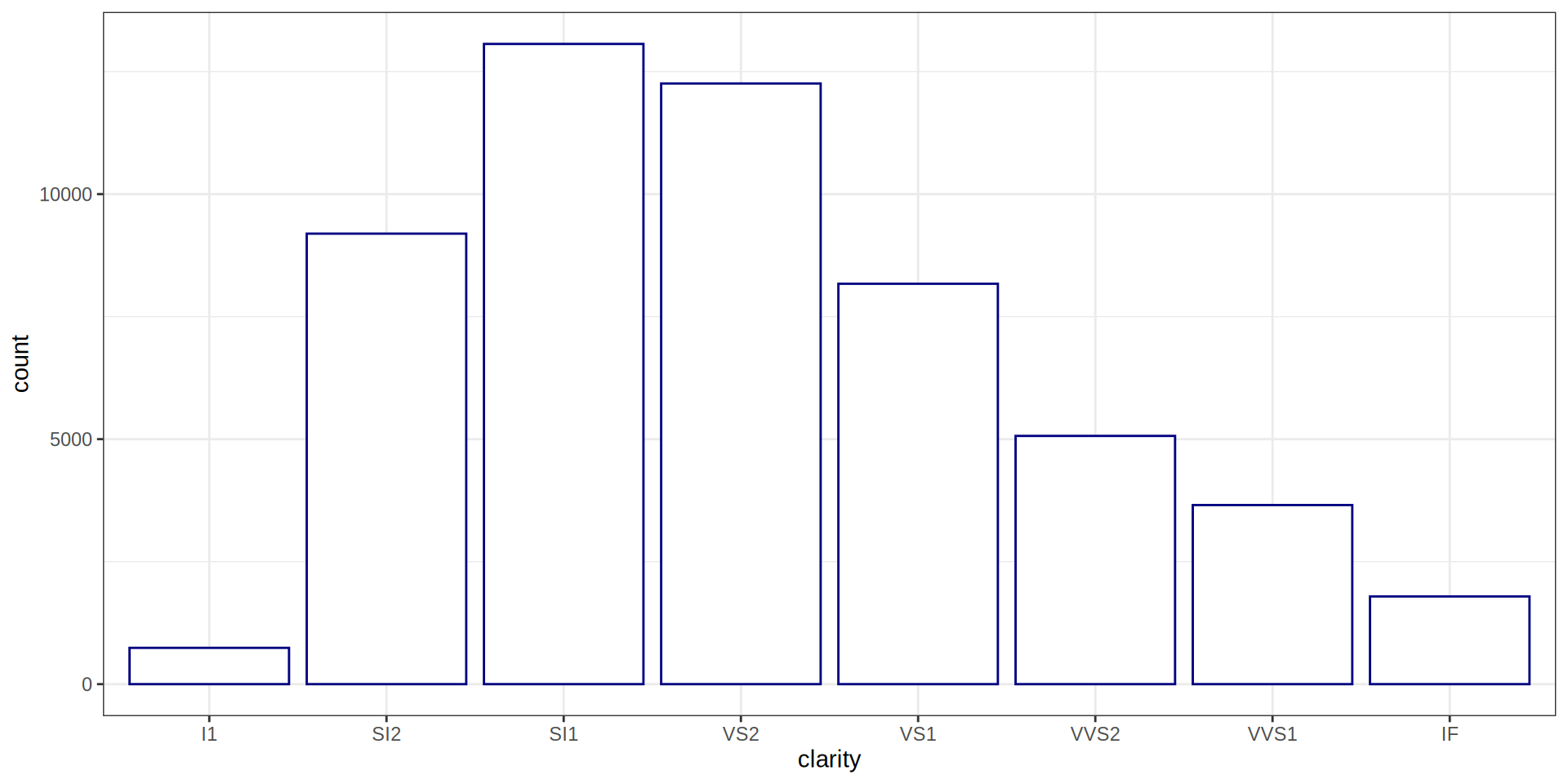
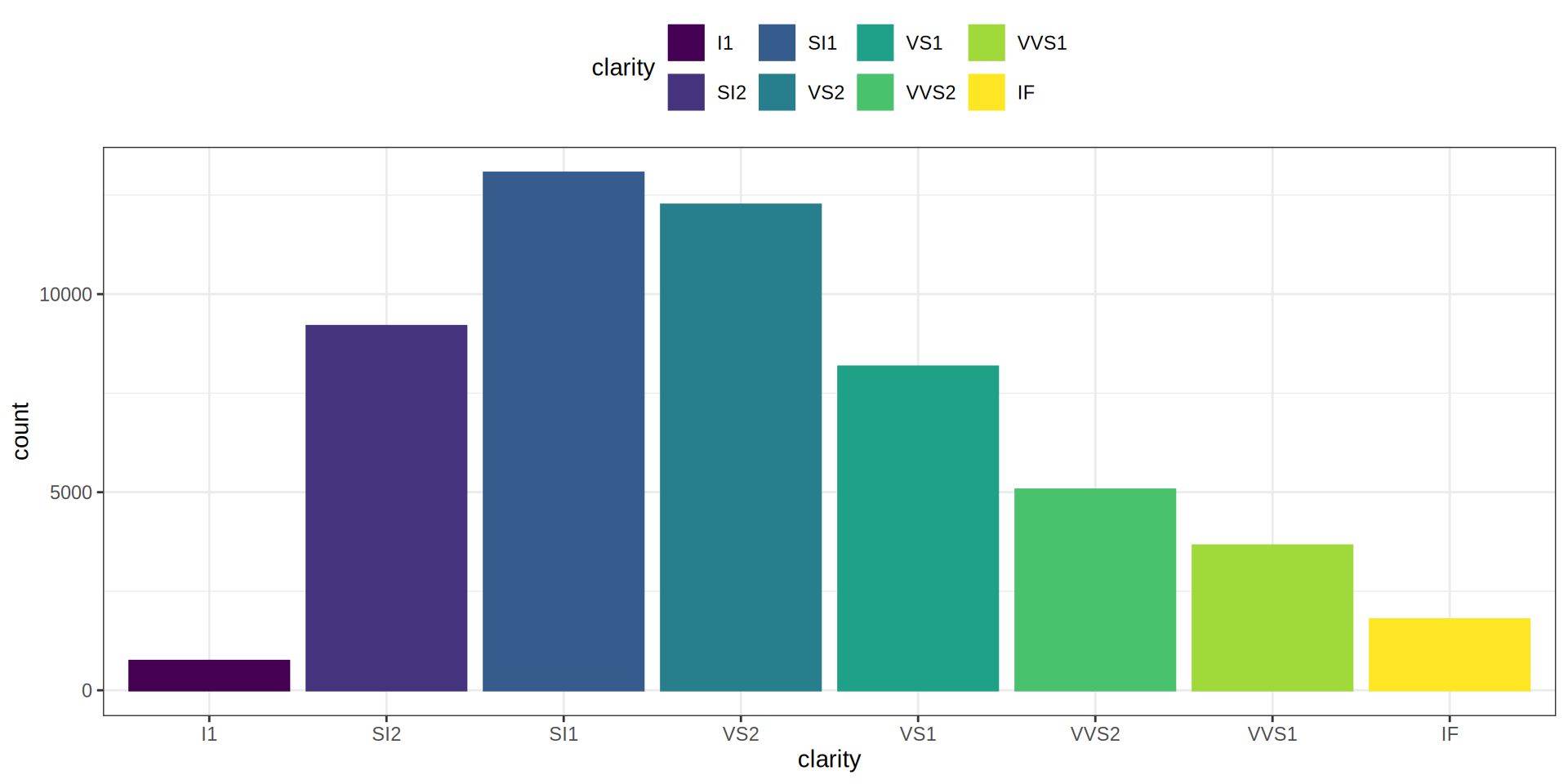
carat cut color clarity depth
Min. :0.2000 Fair : 1610 D: 6775 SI1 :13065 Min. :43.00
1st Qu.:0.4000 Good : 4906 E: 9797 VS2 :12258 1st Qu.:61.00
Median :0.7000 Very Good:12082 F: 9542 SI2 : 9194 Median :61.80
Mean :0.7979 Premium :13791 G:11292 VS1 : 8171 Mean :61.75
3rd Qu.:1.0400 Ideal :21551 H: 8304 VVS2 : 5066 3rd Qu.:62.50
Max. :5.0100 I: 5422 VVS1 : 3655 Max. :79.00
J: 2808 (Other): 2531
table price x y
Min. :43.00 Min. : 326 Min. : 0.000 Min. : 0.000
1st Qu.:56.00 1st Qu.: 950 1st Qu.: 4.710 1st Qu.: 4.720
Median :57.00 Median : 2401 Median : 5.700 Median : 5.710
Mean :57.46 Mean : 3933 Mean : 5.731 Mean : 5.735
3rd Qu.:59.00 3rd Qu.: 5324 3rd Qu.: 6.540 3rd Qu.: 6.540
Max. :95.00 Max. :18823 Max. :10.740 Max. :58.900
z
Min. : 0.000
1st Qu.: 2.910
Median : 3.530
Mean : 3.539
3rd Qu.: 4.040
Max. :31.800
Scatterplot
Vamos a representar carat (quilates) vs price
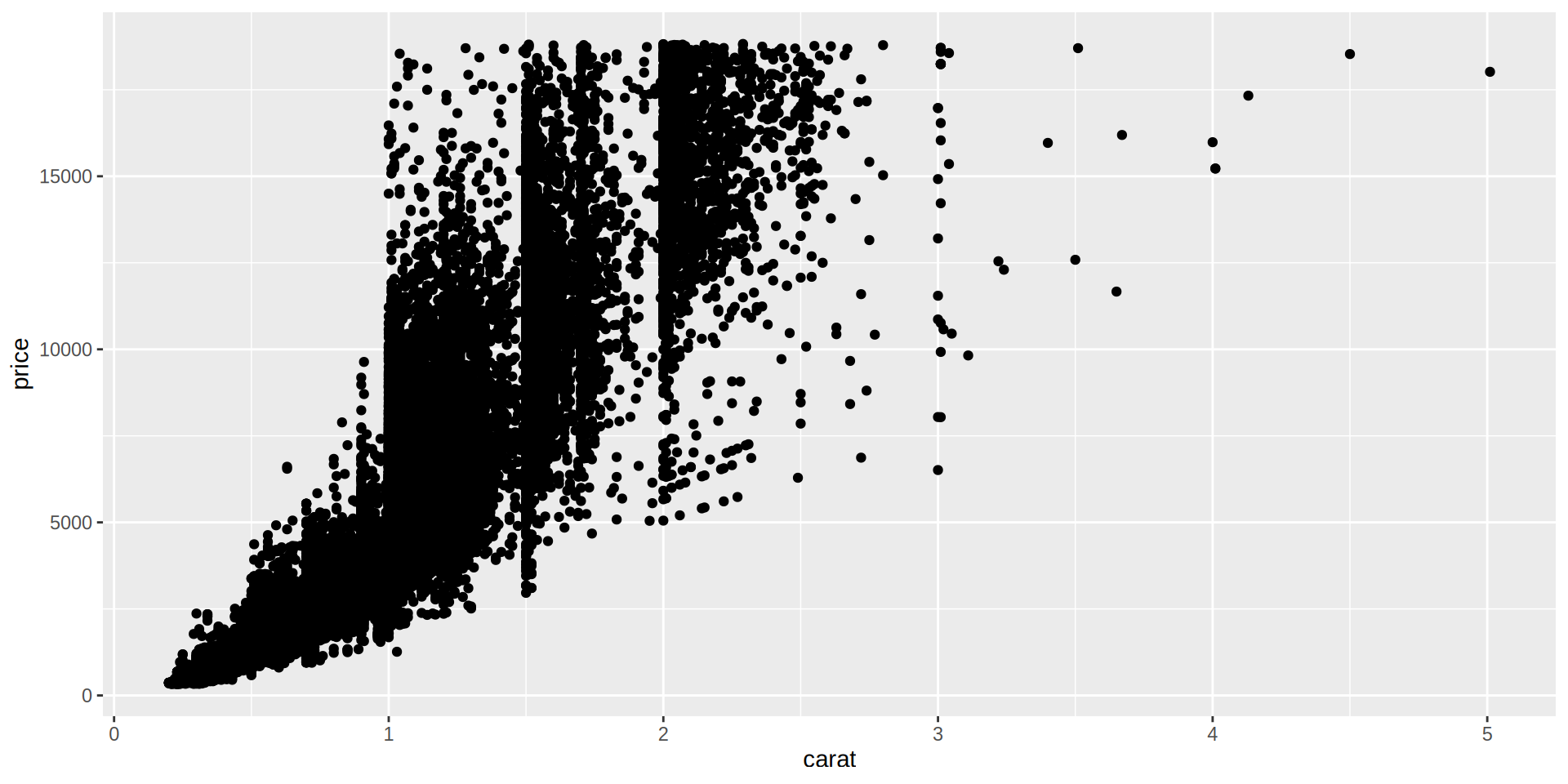
Scatterplot
Añadimos un tema y cambiamos el color de los puntos

Scatterplot
Coloreamos los puntos agrupados por la variable cut. Además movemos la leyenda a la parte superior
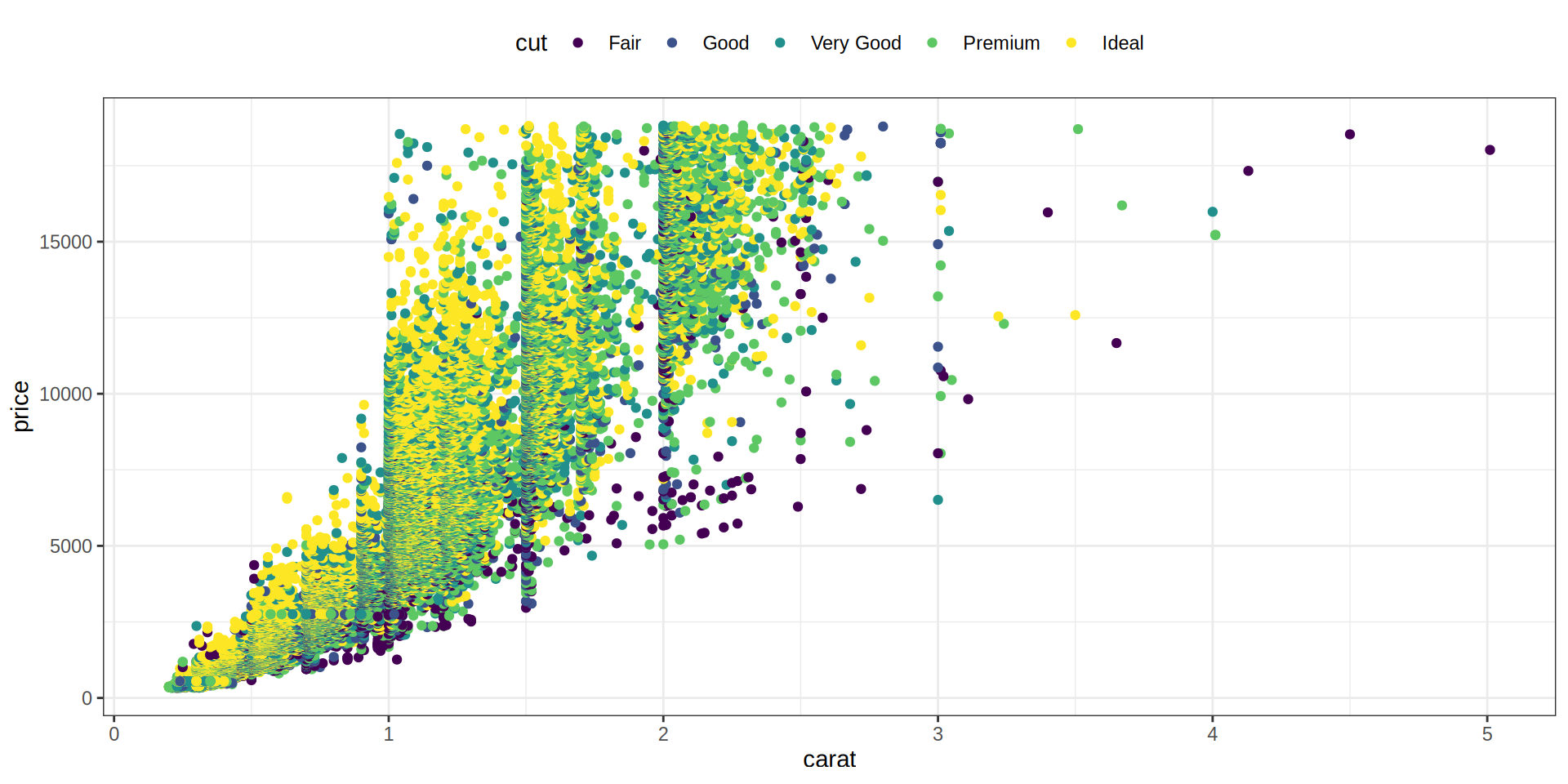
Scatterplot

Boxplot
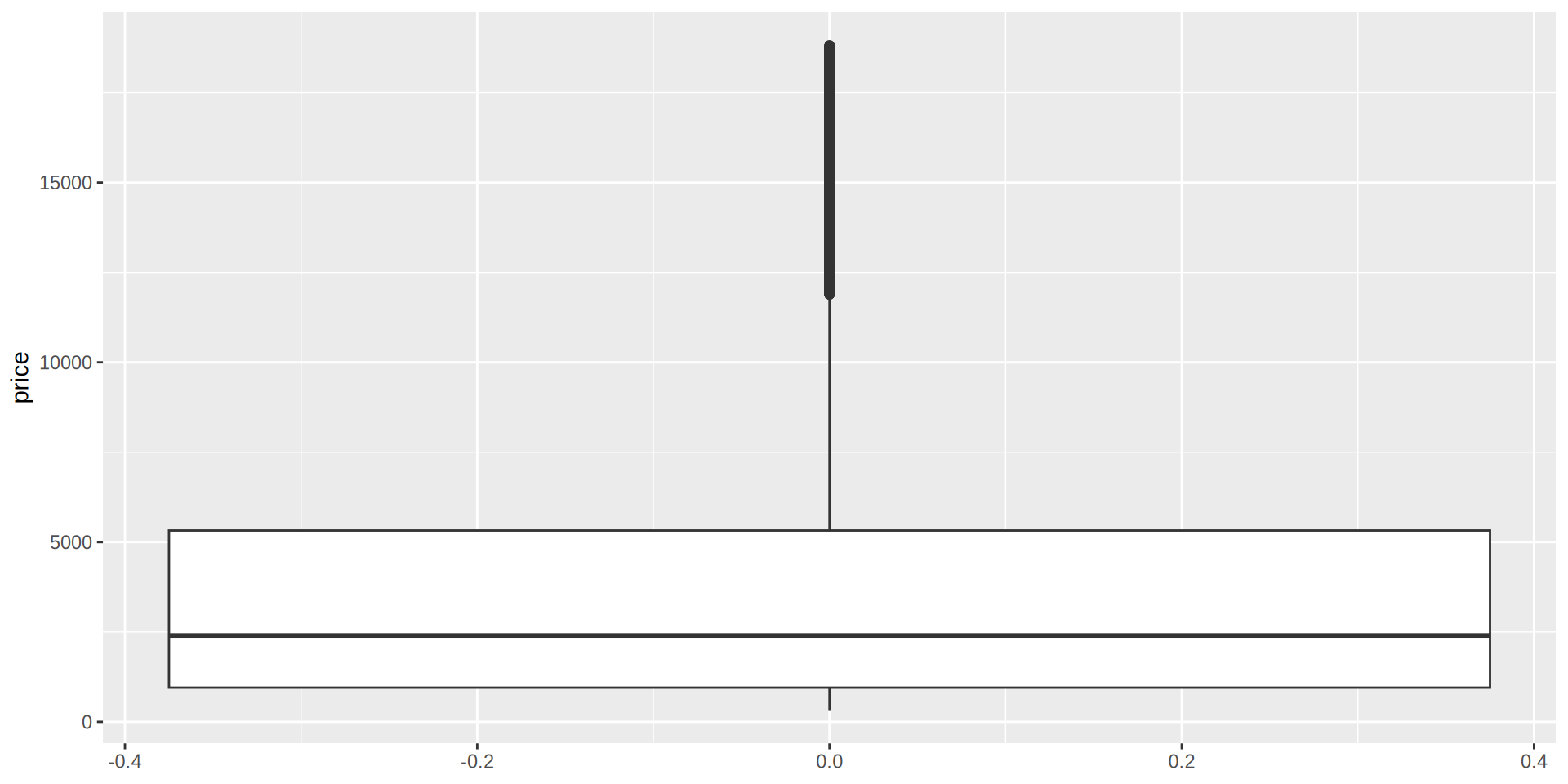
Boxplot
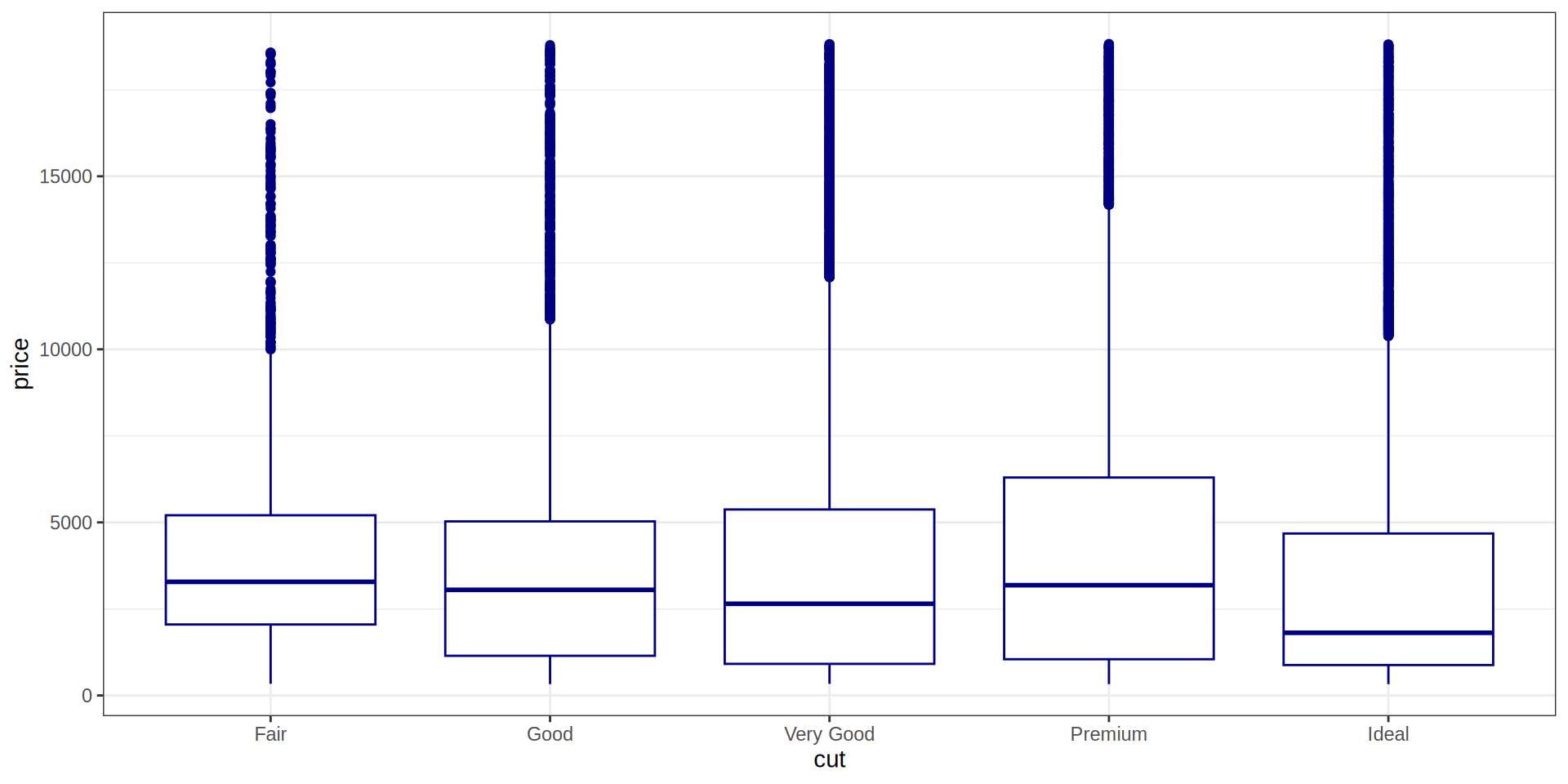
Boxplot
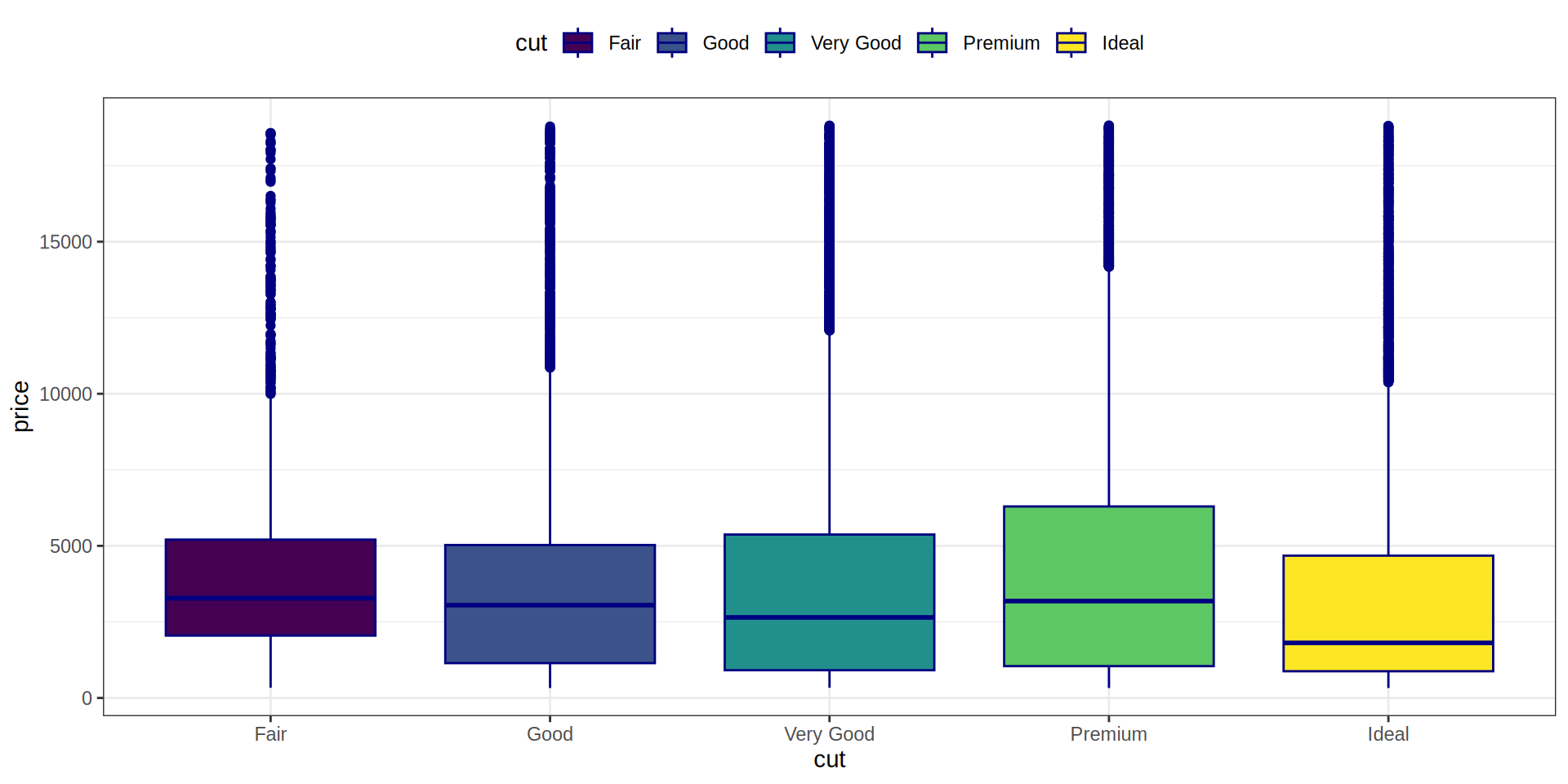
Boxplot
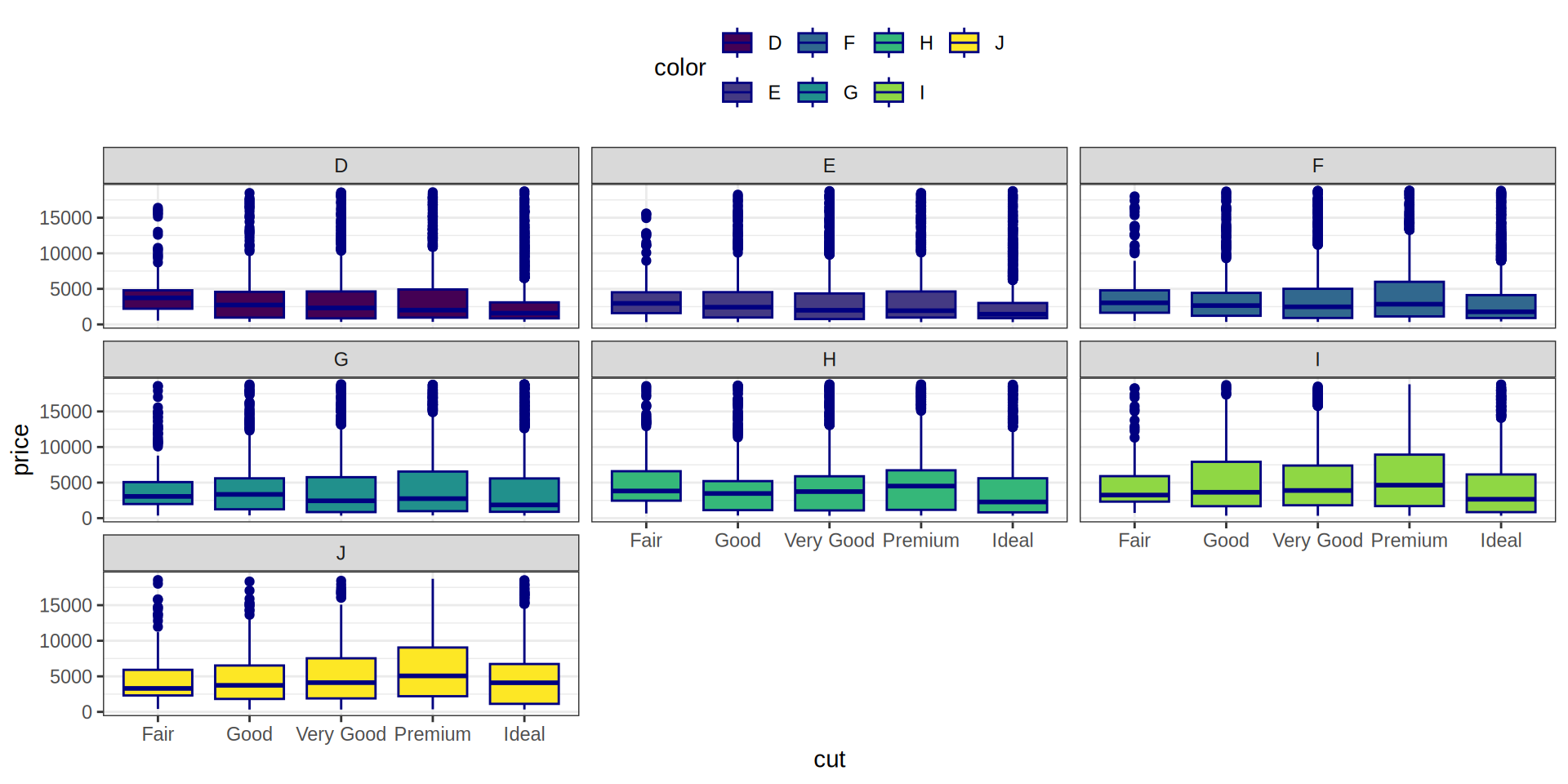
Añadimos una 3ª variable, la variable ‘color’
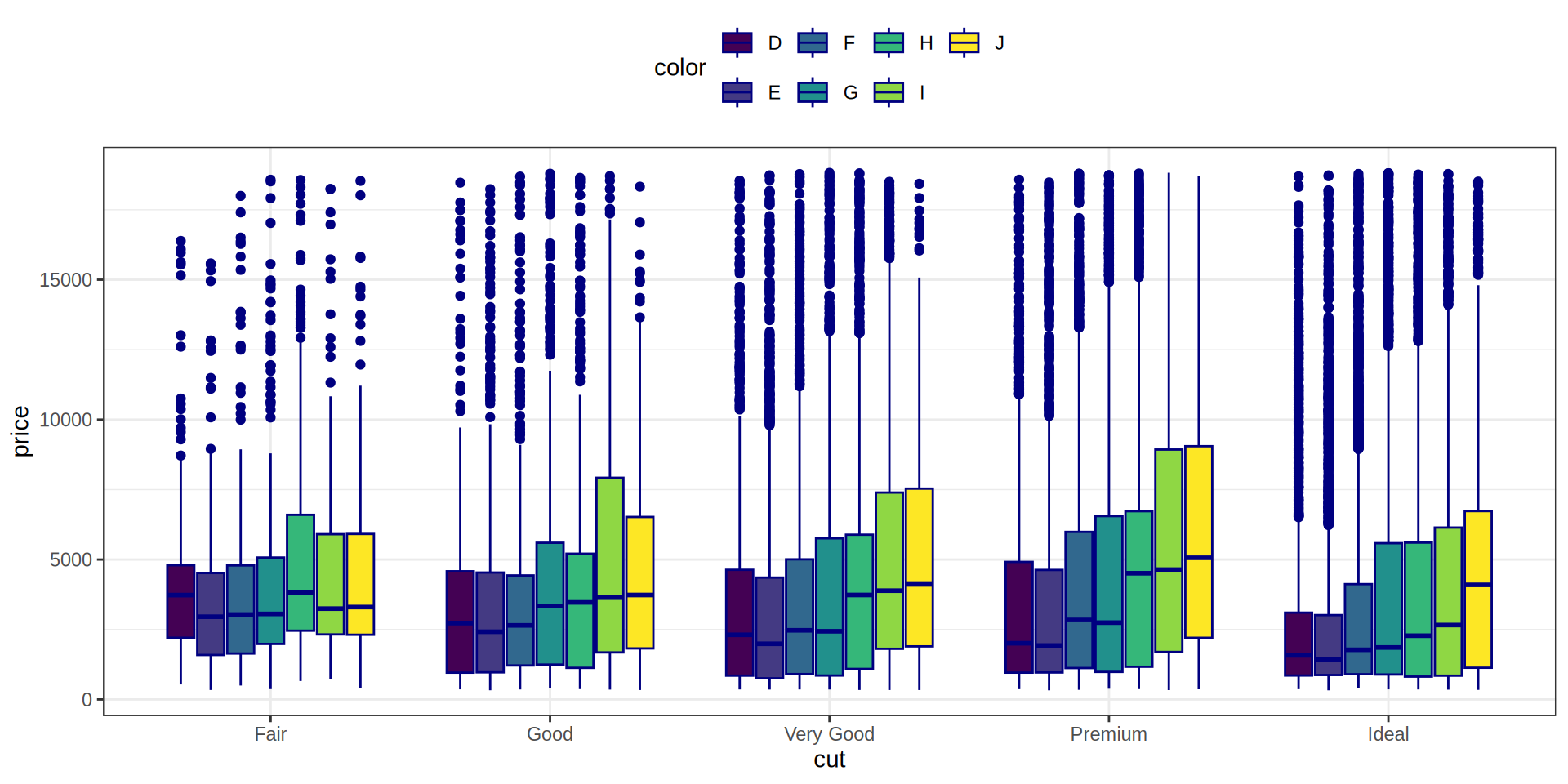
Boxplot
Histogram
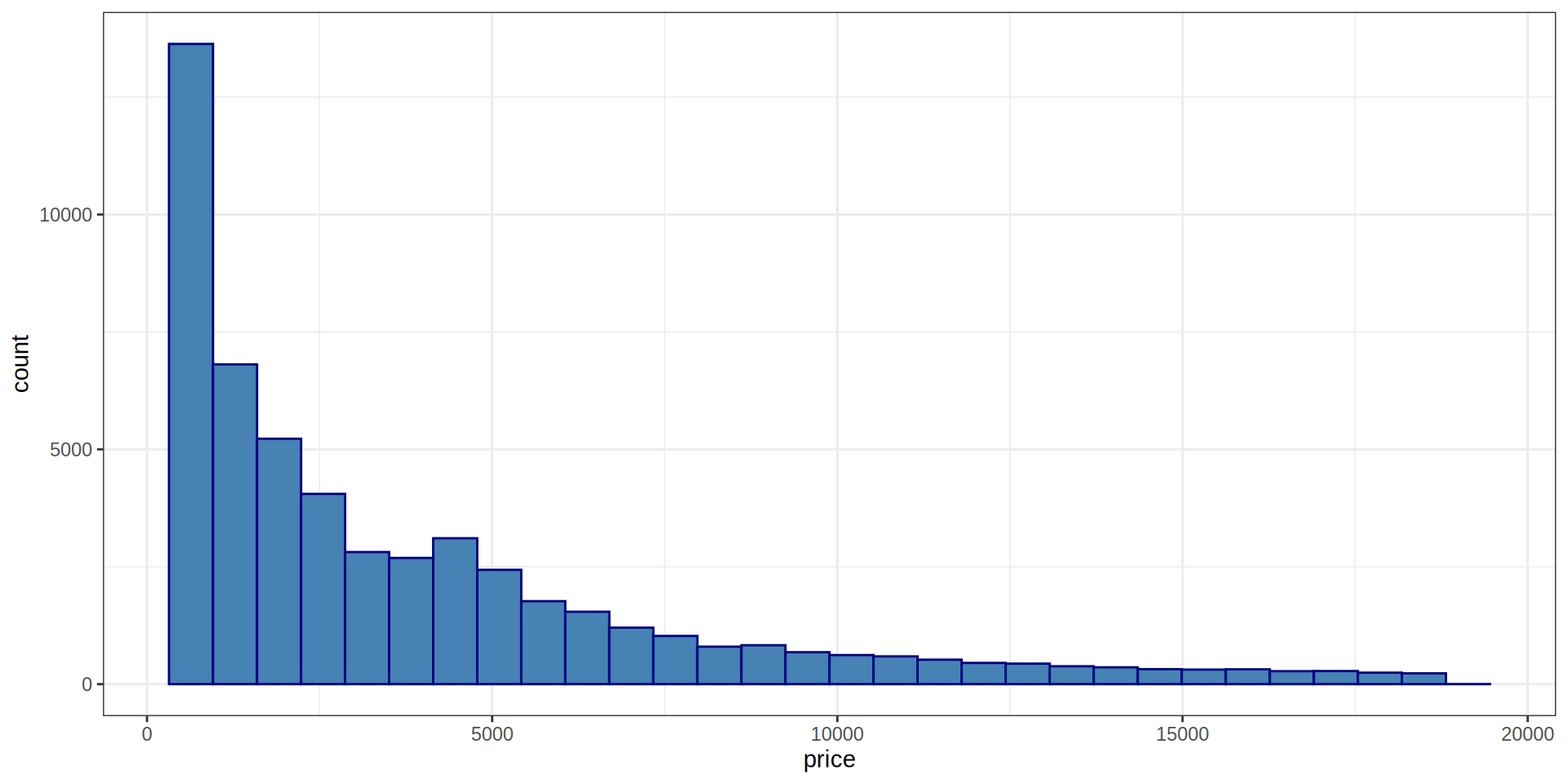
Histogram
Bar Chart
Bar Chart
Bar Chart
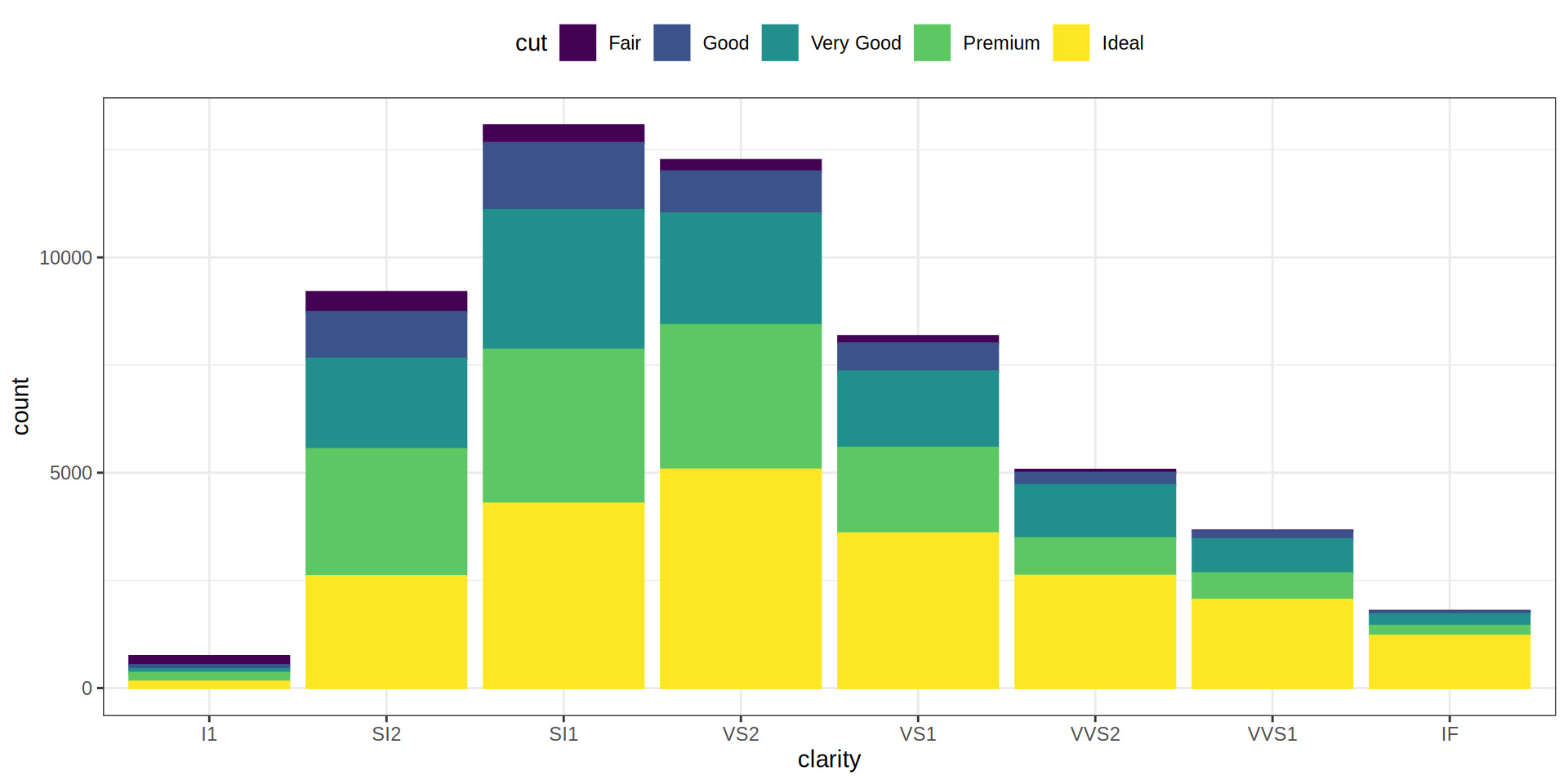
Bar Chart
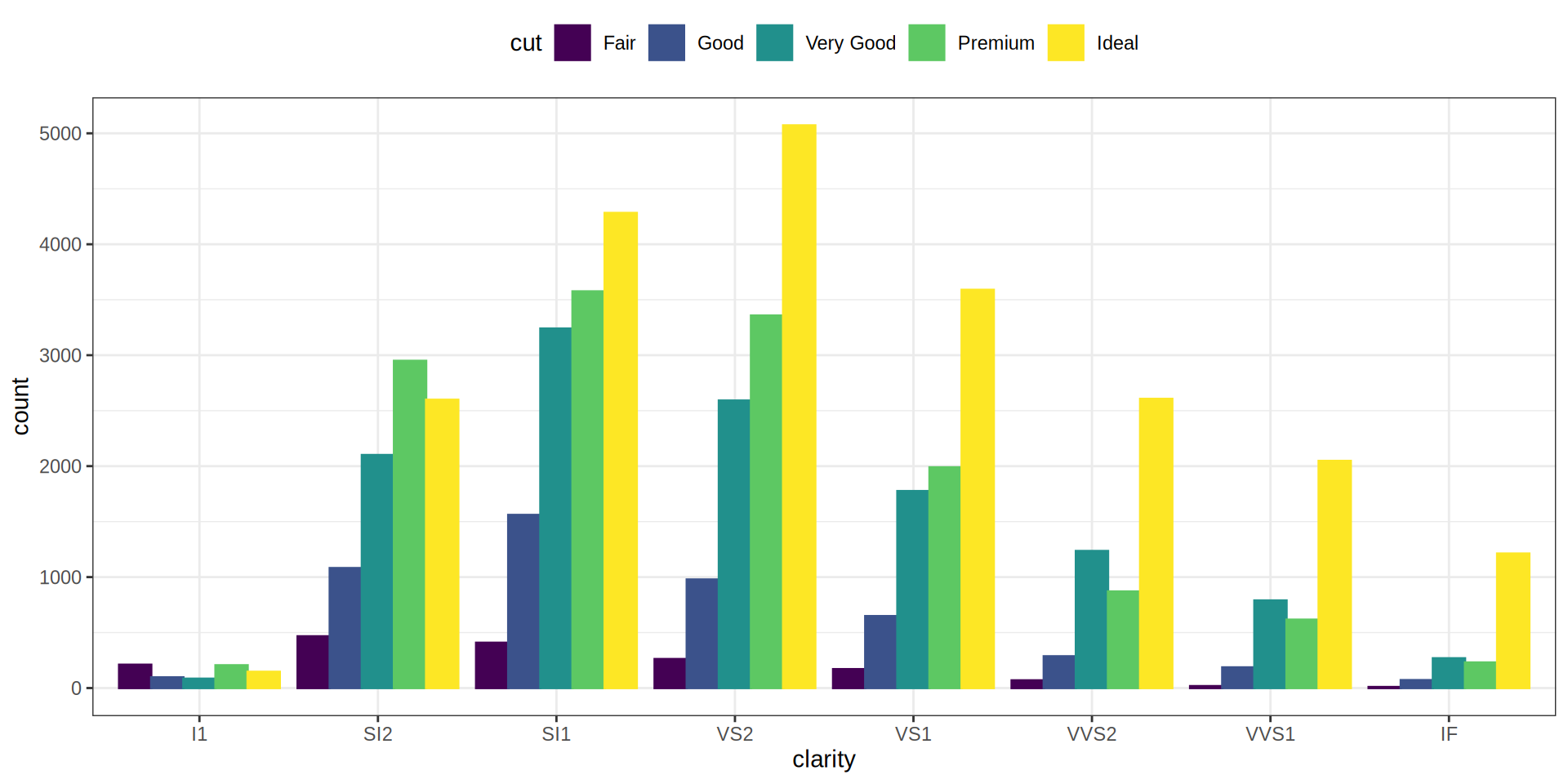
Bar Chart
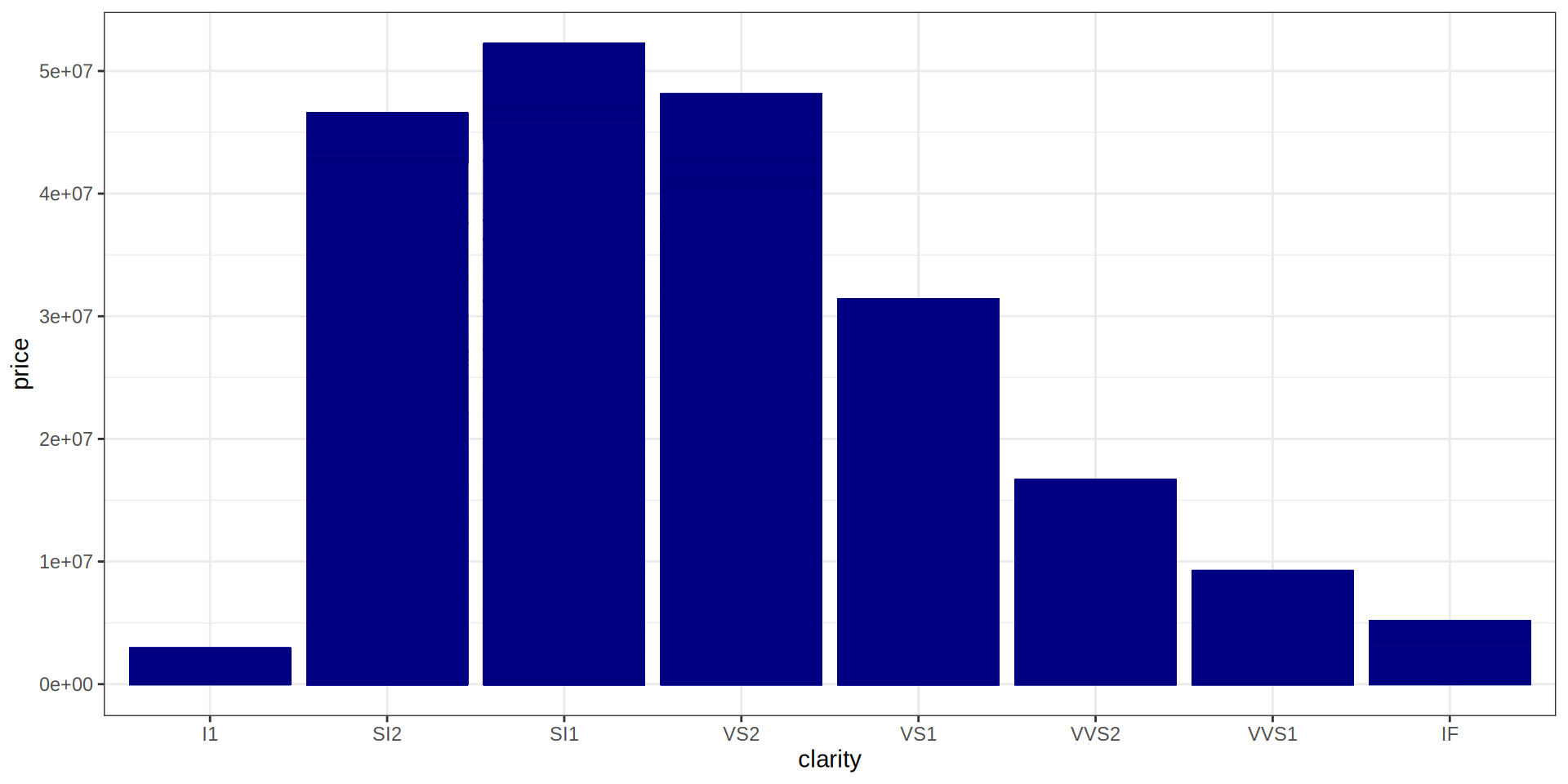
Jitter
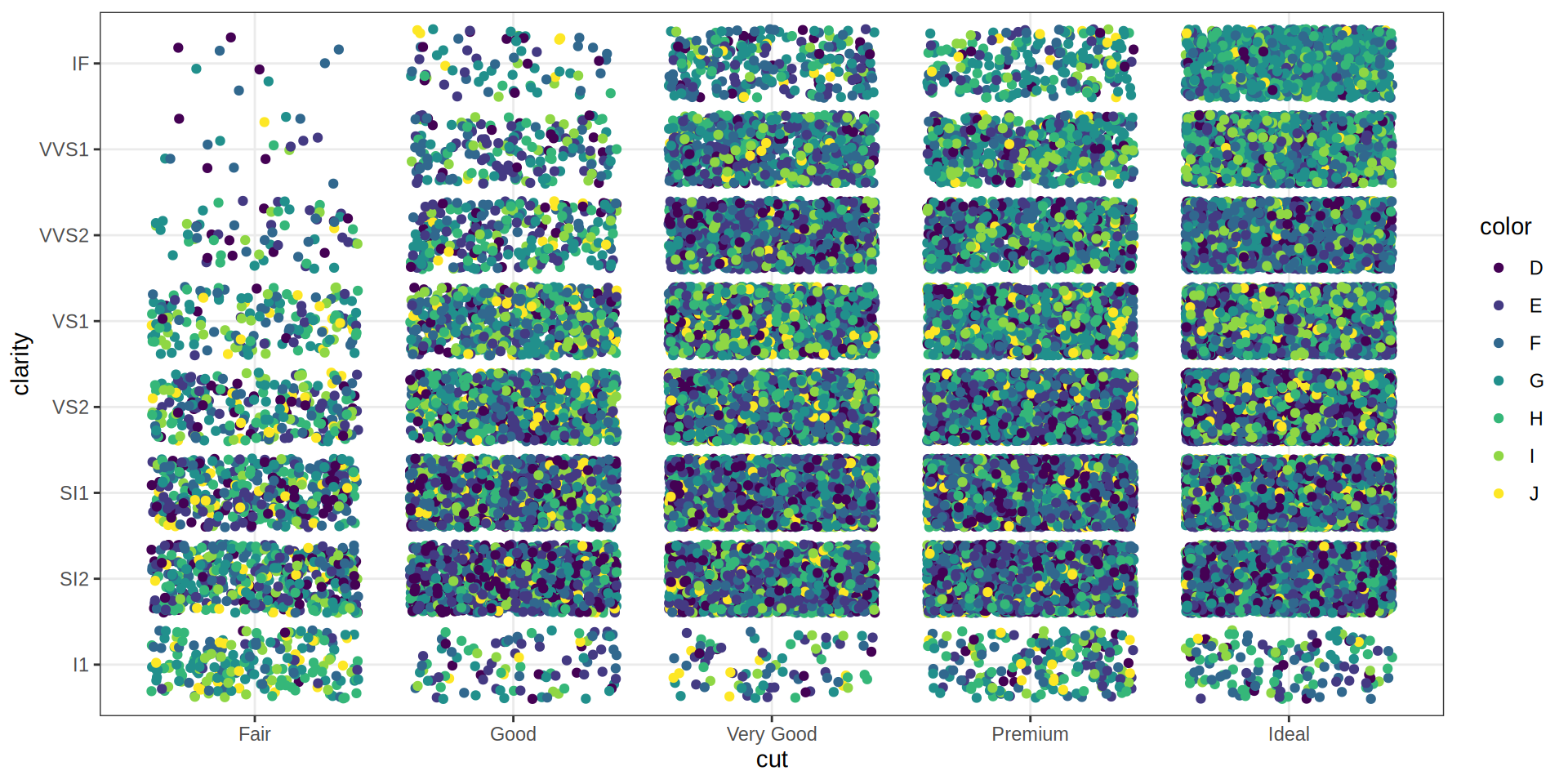
Pie Chart
Volcano Plot
Vamos a realizar el volcano plot correspondiente al análisis diferencial de los datos de Drosophila melanogaster. Eliminamos las filas con 0 o NA values
X baseMean log2FoldChange lfcSE pvalue padj
1 FBgn0000008 562.4671 -0.05656922 0.05473453 3.005379e-01 3.499743e-01
2 FBgn0000014 894.8910 -0.79584159 0.04535355 3.664583e-69 2.568605e-68
3 FBgn0000015 323.3163 -0.84122112 0.07377132 2.177357e-30 8.697926e-30
4 FBgn0000017 862.1217 -0.25306214 0.04781241 1.098852e-07 2.259261e-07
5 FBgn0000018 111.7811 0.37605333 0.11157453 6.718636e-04 1.085105e-03
6 FBgn0000024 789.7766 -0.91597718 0.04844234 5.862028e-80 4.626392e-79Volcano Plot
Seleccionamos los valores de FC = 2 y padj = 0.0000001 de corte.
X baseMean log2FoldChange lfcSE pvalue padj
1 FBgn0000008 562.4671 -0.05656922 0.05473453 3.005379e-01 3.499743e-01
2 FBgn0000014 894.8910 -0.79584159 0.04535355 3.664583e-69 2.568605e-68
3 FBgn0000015 323.3163 -0.84122112 0.07377132 2.177357e-30 8.697926e-30
4 FBgn0000017 862.1217 -0.25306214 0.04781241 1.098852e-07 2.259261e-07
5 FBgn0000018 111.7811 0.37605333 0.11157453 6.718636e-04 1.085105e-03
6 FBgn0000024 789.7766 -0.91597718 0.04844234 5.862028e-80 4.626392e-79
class
1 none
2 none
3 none
4 none
5 none
6 noneVolcano Plot
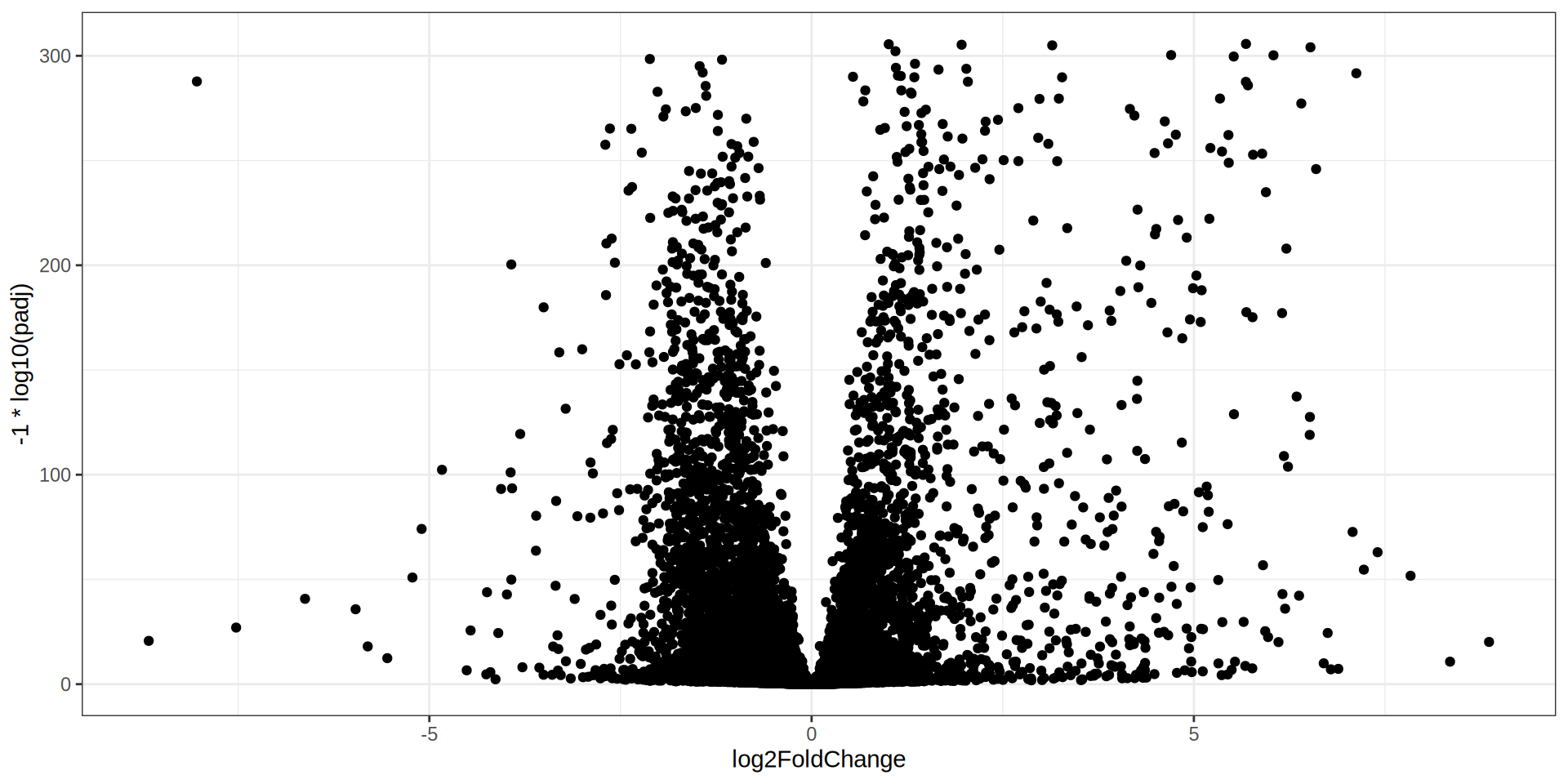
Volcano Plot
ggplot(dge, aes(x = log2FoldChange, y = -1 * log10(padj))) +
geom_point() +
geom_hline(yintercept = -1 * log10(padj.cutoff ), linetype="dashed",
color = "black", linewidth = 0.2) +
geom_vline(xintercept=c(-1 * lfc.cutoff ,lfc.cutoff ), linetype="dashed",
color = "black", linewidth = 0.2) +
theme_bw()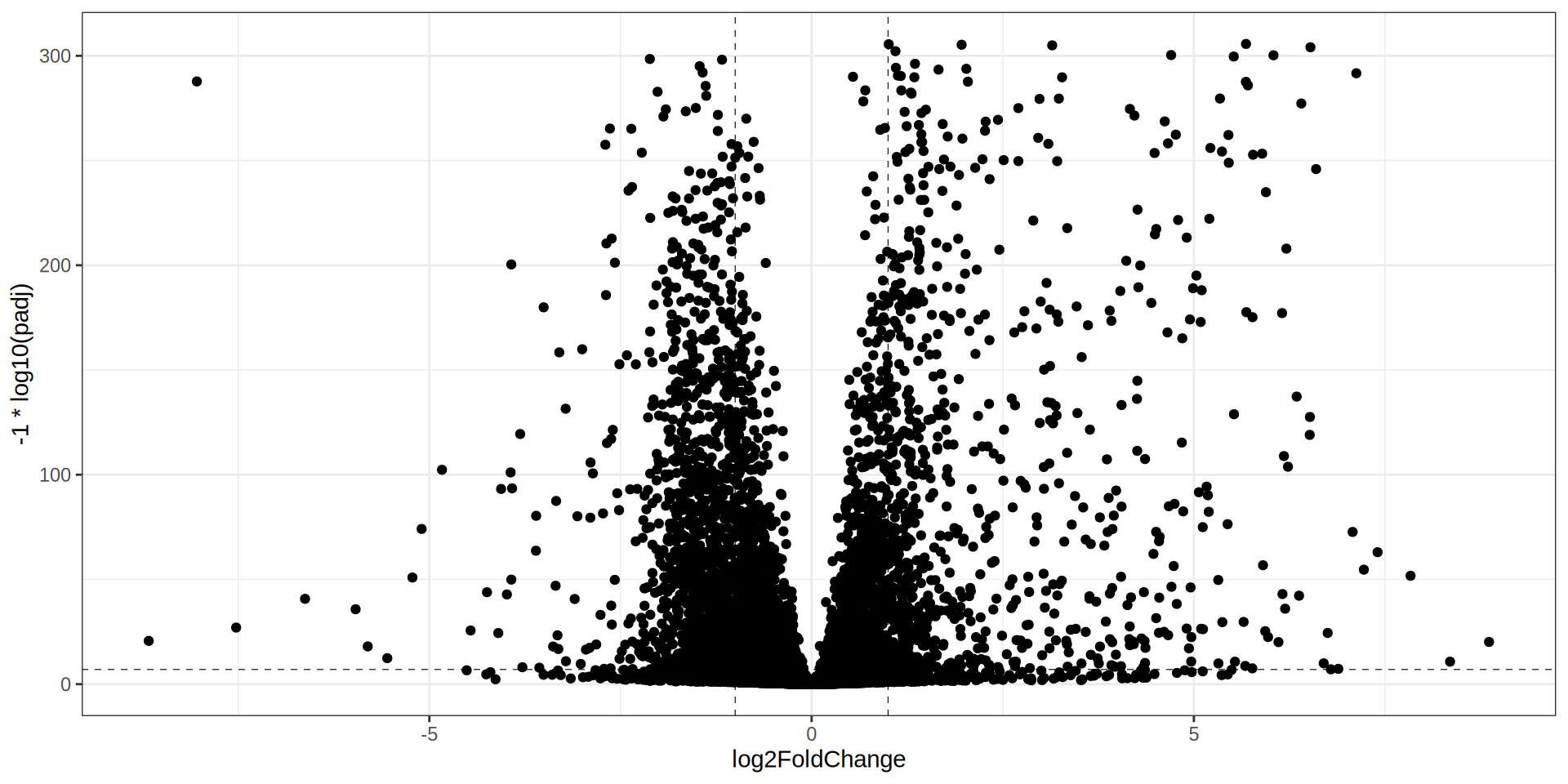
Volcano Plot
ggplot(dge, aes(x = log2FoldChange, y = -1 * log10(padj))) +
geom_point(aes(color = class)) +
geom_hline(yintercept = -1 * log10(padj.cutoff ), linetype="dashed",
color = "black", linewidth = 0.2) +
geom_vline(xintercept=c(-1 * lfc.cutoff ,lfc.cutoff ), linetype="dashed",
color = "black", linewidth = 0.2) +
theme_bw()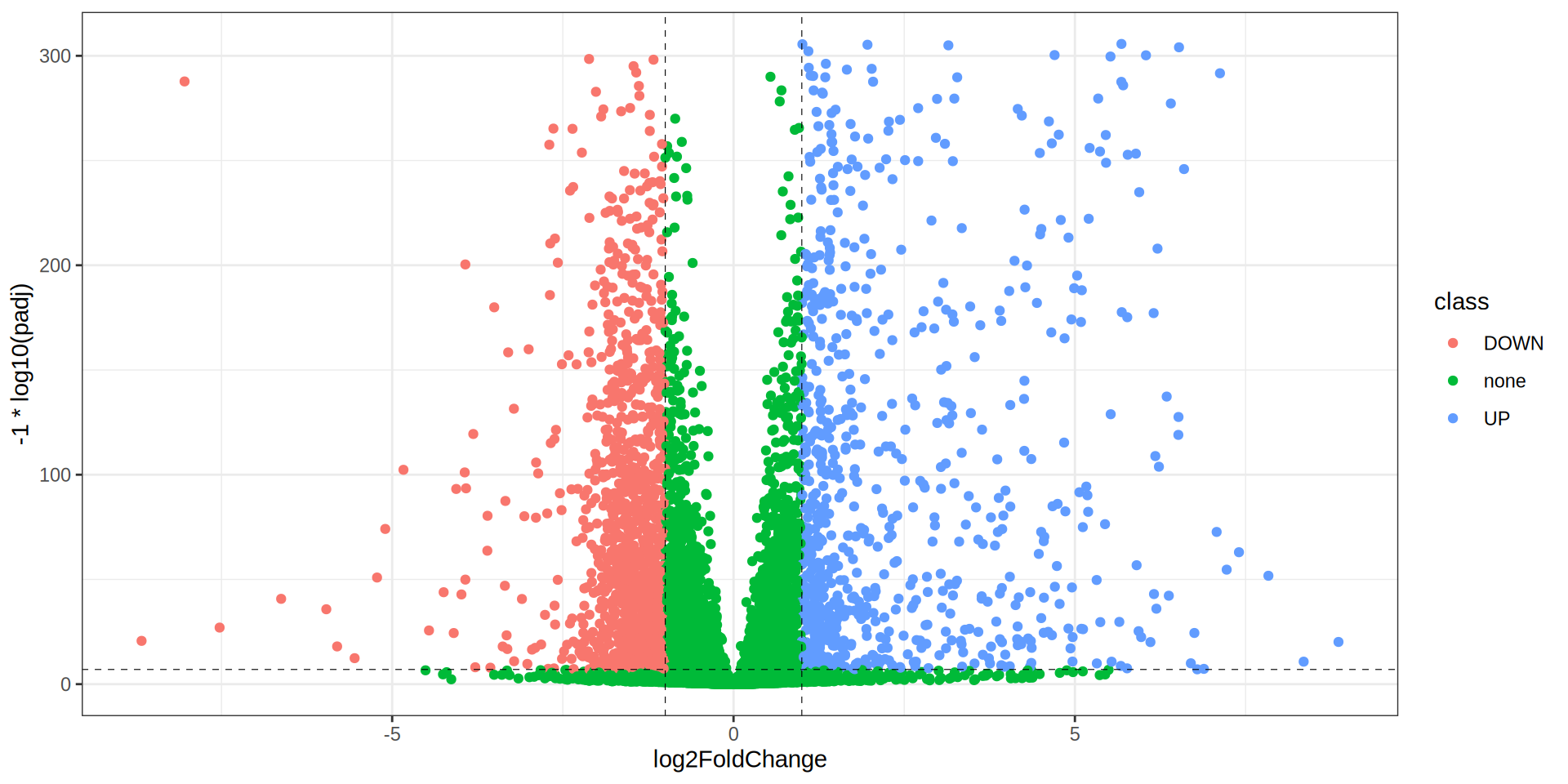
Volcano Plot
colors <- c("UP"="#FC4E07", "none"="#E7B800", "DOWN"="#00AFBB")
ggplot(dge, aes(x = log2FoldChange, y = -1 * log10(padj))) +
geom_point(aes(color = class)) +
scale_color_manual(values = colors) +
geom_hline(yintercept = -1 * log10(padj.cutoff ), linetype="dashed",
color = "black", linewidth = 0.2) +
geom_vline(xintercept=c(-1 * lfc.cutoff ,lfc.cutoff ), linetype="dashed",
color = "black", linewidth = 0.2) +
theme_bw()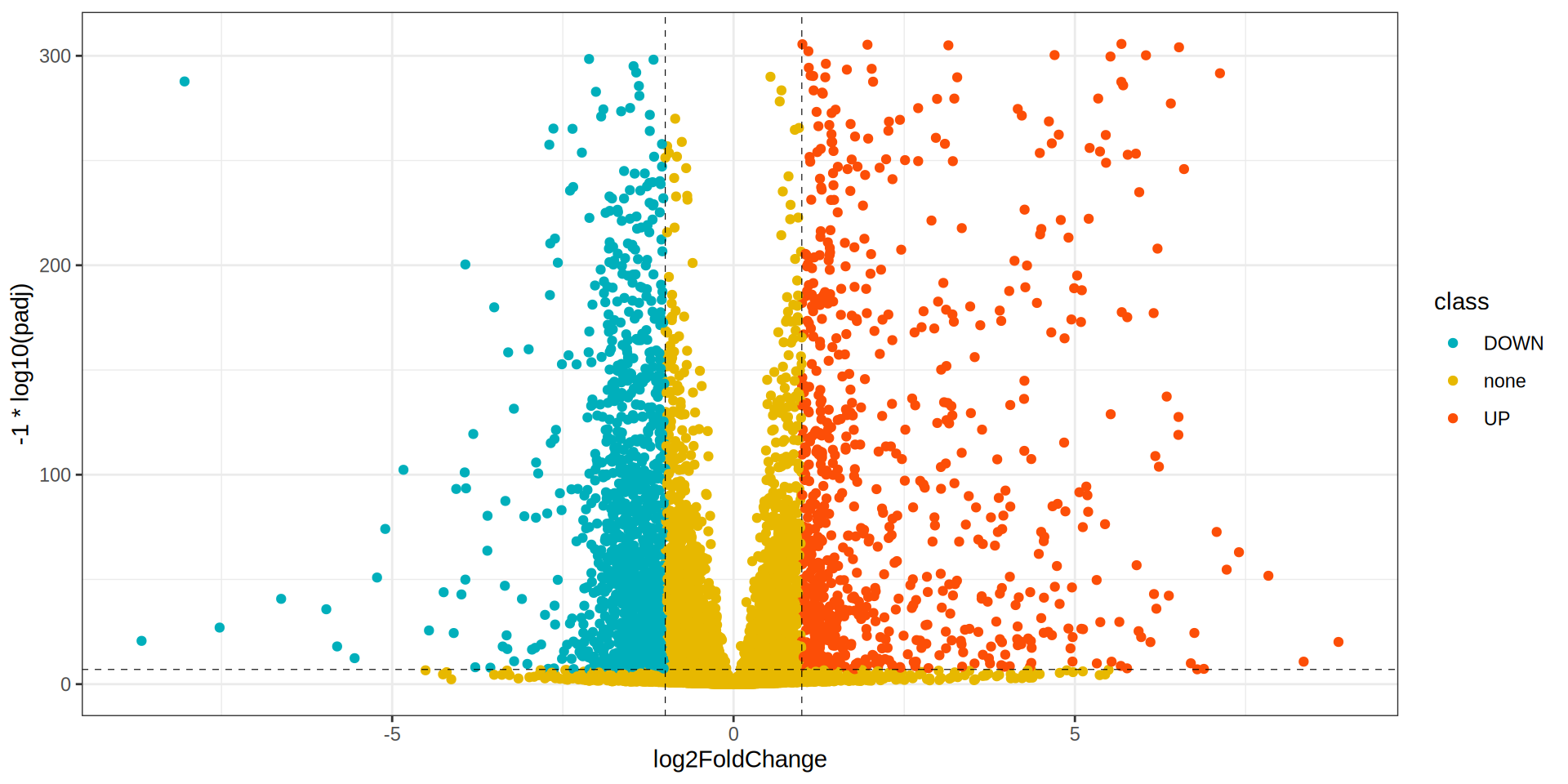
Volcano Plot
library(ggrepel)
ggplot(dge, aes(x = log2FoldChange, y = -1 * log10(padj), label = X)) +
geom_point(aes(color = class)) +
scale_color_manual(values = colors) +
geom_hline(yintercept = -1 * log10(padj.cutoff ), linetype="dashed",
color = "black", linewidth = 0.2) +
geom_vline(xintercept=c(-1 * lfc.cutoff ,lfc.cutoff ), linetype="dashed",
color = "black", linewidth = 0.2) +
geom_label_repel(data = dge[dge$class %in% c('UP', 'DOWN'), ],
size= 4, color = 'firebrick', point.padding=unit(0.5, "lines"), max.overlaps = 30) +
theme_bw()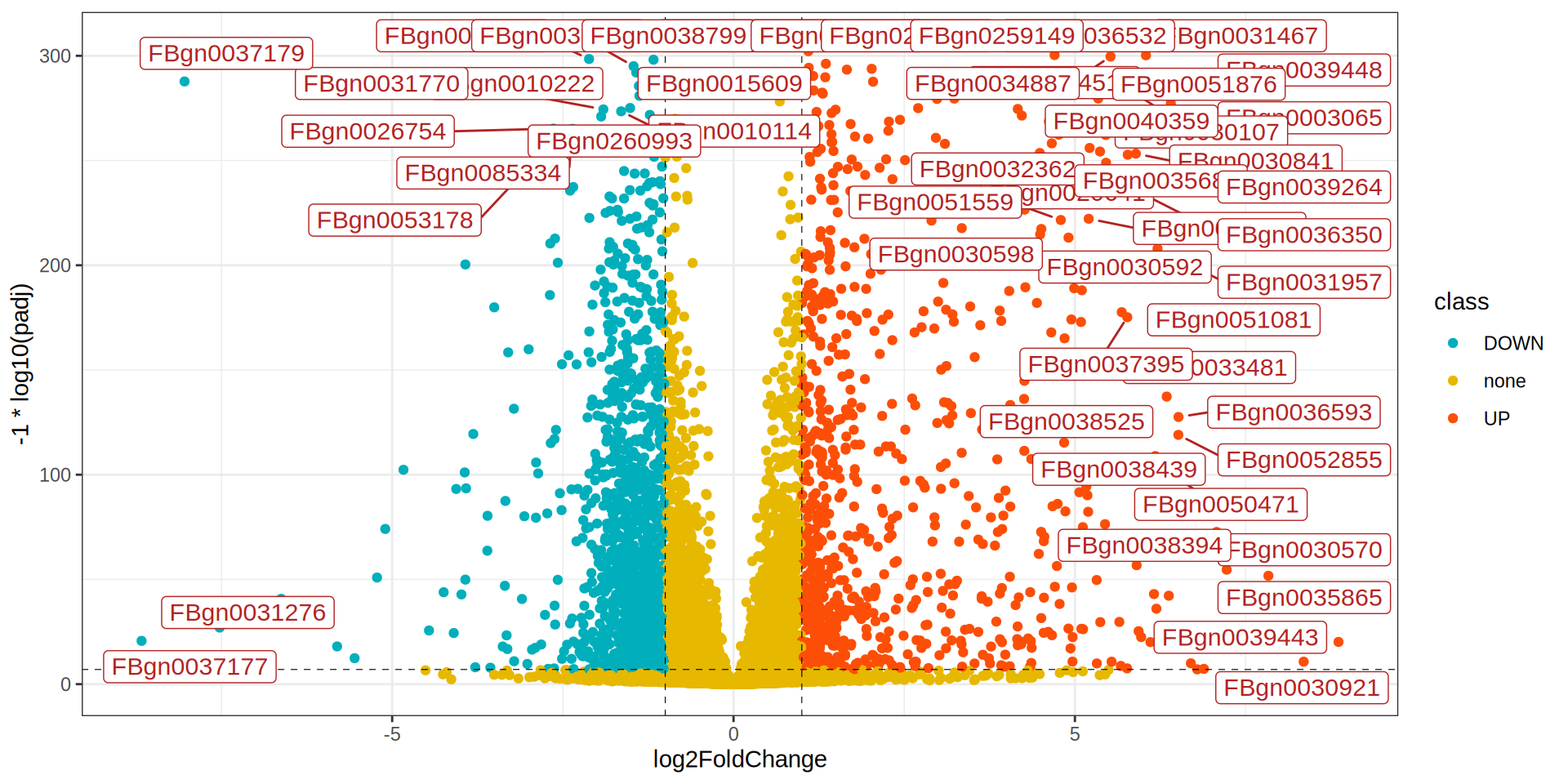
Volcano Plot
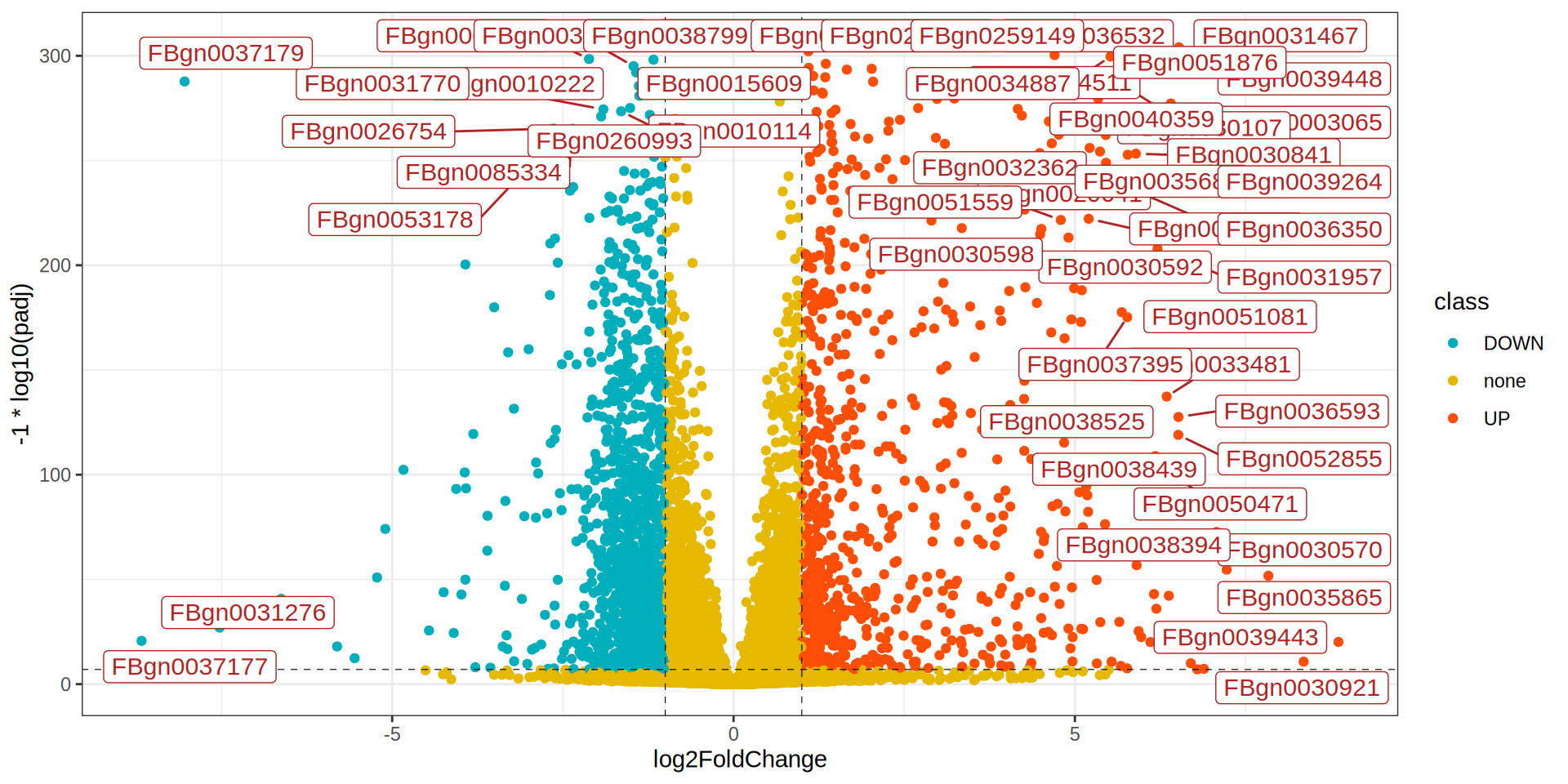
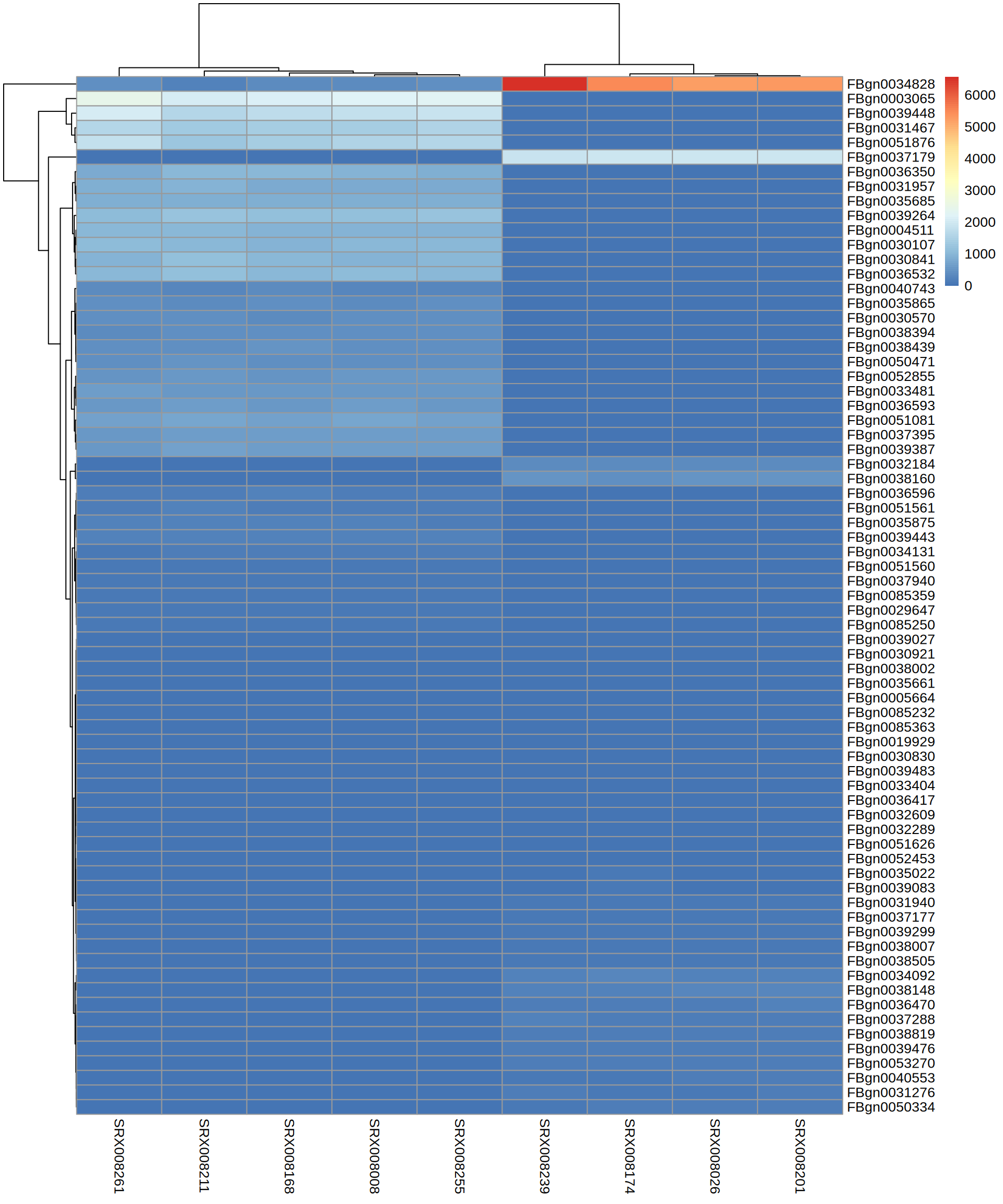
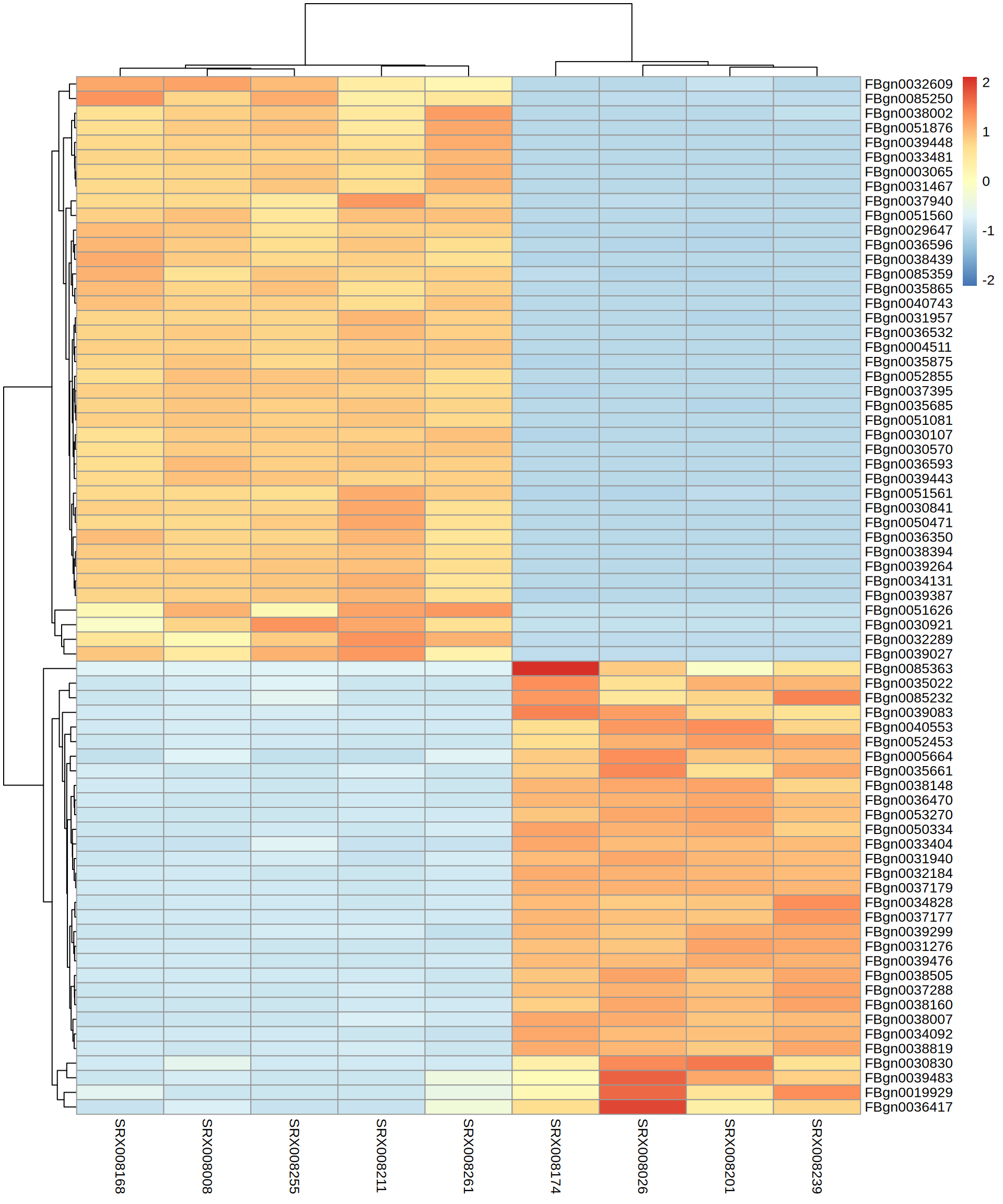
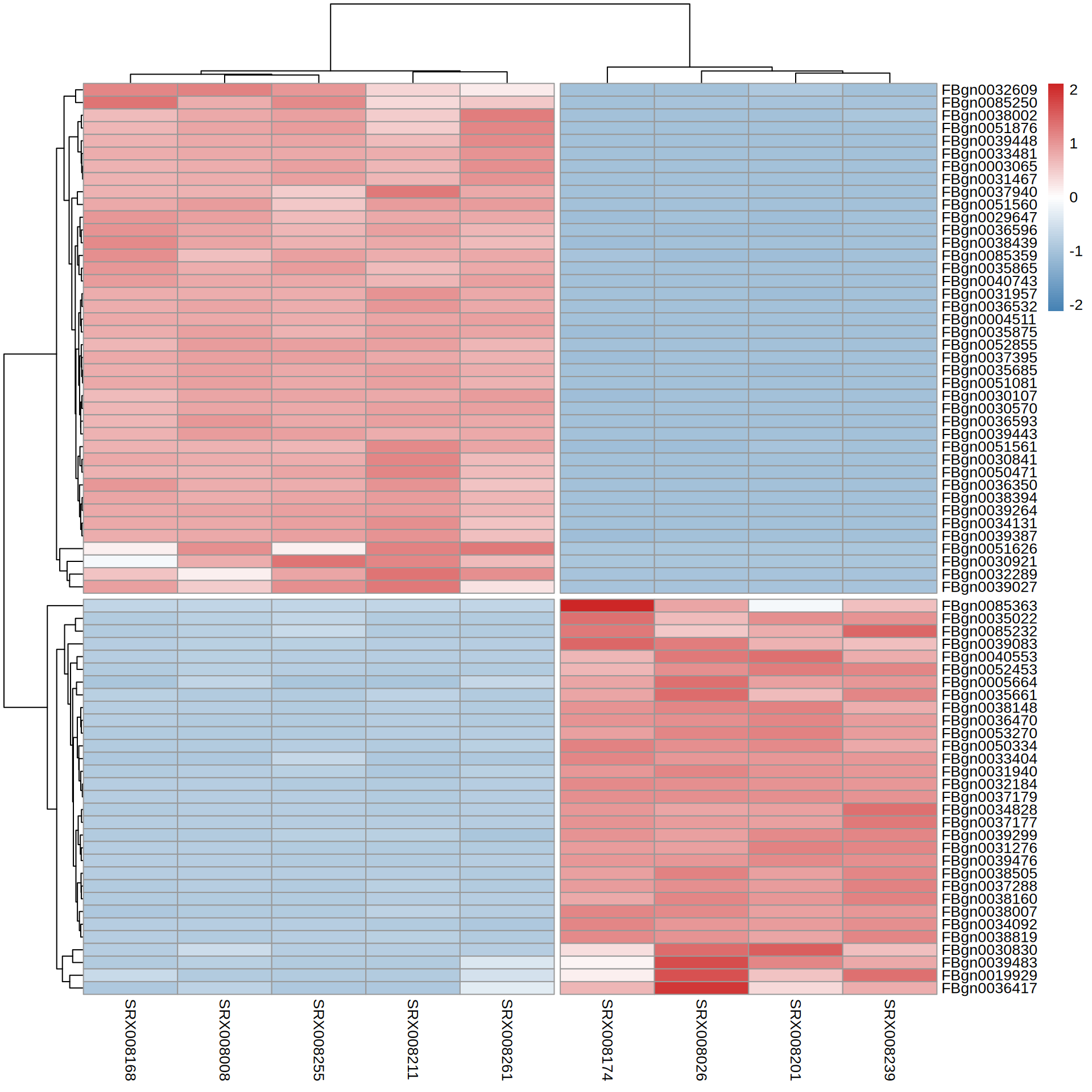
TOP Genes
[1] "FBgn0039443" "FBgn0030921" "FBgn0035865" "FBgn0030570" "FBgn0040743"
[6] "FBgn0039448" "FBgn0038394" "FBgn0032289" "FBgn0051626" "FBgn0051560"
[11] "FBgn0039027" "FBgn0039264" "FBgn0031467" "FBgn0036593" "FBgn0052855"
[16] "FBgn0003065" "FBgn0036596" "FBgn0033481" "FBgn0050471" "FBgn0031957"
[21] "FBgn0034131" "FBgn0038439" "FBgn0051561" "FBgn0051081" "FBgn0085359"
[26] "FBgn0051876" "FBgn0037940" "FBgn0036350" "FBgn0085250" "FBgn0035875"
[31] "FBgn0030841" "FBgn0035685" "FBgn0037395" "FBgn0032609" "FBgn0030107"
[36] "FBgn0039387" "FBgn0036532" "FBgn0004511" "FBgn0038002" "FBgn0029647" [1] "FBgn0036417" "FBgn0032184" "FBgn0030830" "FBgn0005664" "FBgn0039476"
[6] "FBgn0050334" "FBgn0085232" "FBgn0034092" "FBgn0037288" "FBgn0038007"
[11] "FBgn0038160" "FBgn0034828" "FBgn0031940" "FBgn0036470" "FBgn0039083"
[16] "FBgn0085363" "FBgn0039483" "FBgn0039299" "FBgn0033404" "FBgn0035022"
[21] "FBgn0019929" "FBgn0038148" "FBgn0038819" "FBgn0040553" "FBgn0035661"
[26] "FBgn0052453" "FBgn0038505" "FBgn0053270" "FBgn0031276" "FBgn0037179"
[31] "FBgn0037177"TOP Genes
X SRX008026 SRX008174 SRX008201 SRX008239 SRX008008 SRX008168
1 FBgn0000008 577.30414 589.66391 536.2258 595.58471 560.6214 581.6257
2 FBgn0000014 1147.76523 1197.37875 1121.8777 1217.76428 671.0268 695.3146
3 FBgn0000015 395.03031 443.75218 452.2948 431.81641 229.7358 255.3881
4 FBgn0000017 895.81677 1007.84392 952.1506 930.81982 780.1100 792.5268
5 FBgn0000018 93.31425 96.27166 95.1218 97.28117 122.6359 154.8805
6 FBgn0000024 1117.90467 1063.50098 1050.0701 1048.39706 562.2742 561.8537
SRX008211 SRX008255 SRX008261
1 516.9053 536.3239 567.9487
2 682.5334 668.7586 651.6000
3 236.6116 256.0892 209.1284
4 789.9186 771.1942 838.7149
5 120.1259 118.5327 107.8662
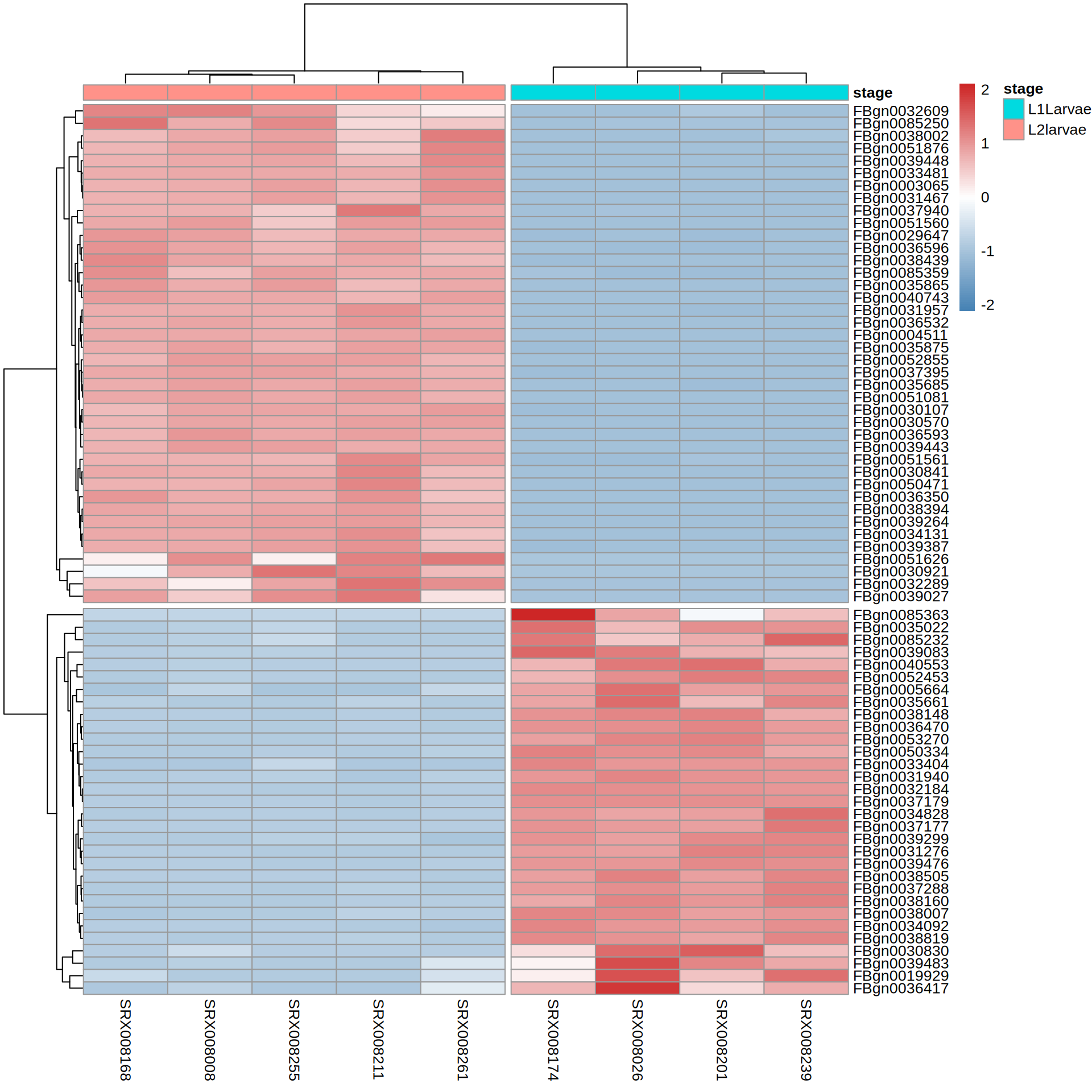
6 607.9097 578.7615 517.3176Heatmap
Heatmap
Heatmap
Heatmap

ggplot2